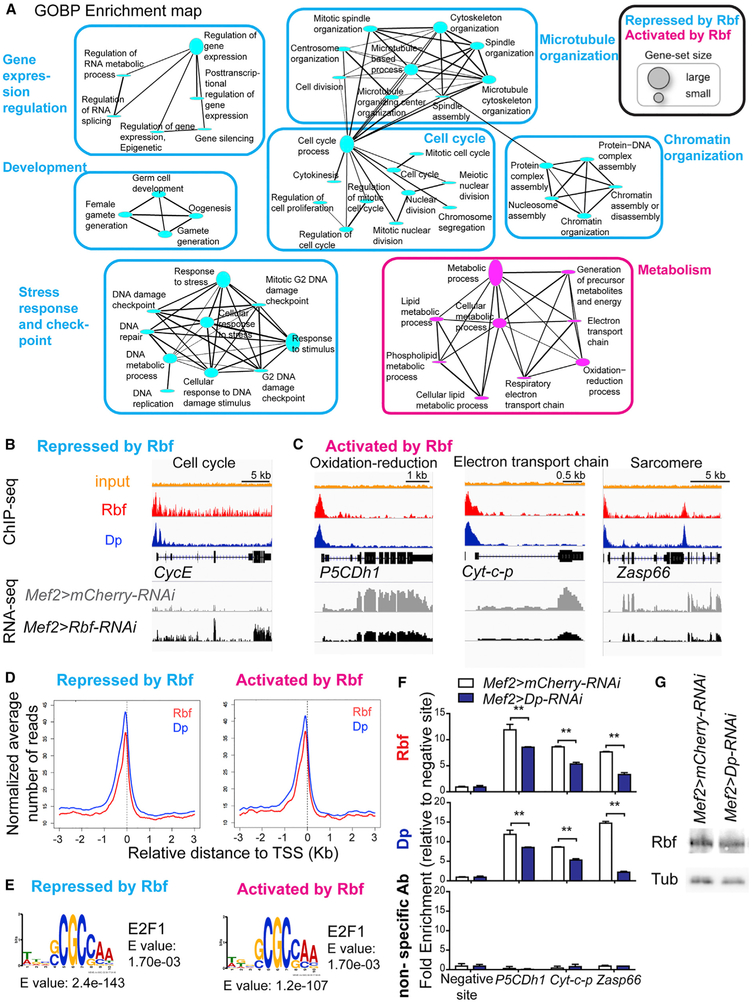
Figure 5. Rbf Activates the Expression of Metabolic Genes in Muscle.
(A) Functional biological processes directly repressed (986 transcripts) and activated (851 transcripts) by Rbf in muscles. GO terms visualized as an enrichment map, clustered using Cytoscape. Size of gene-set is scaled based on the number of genes enriched.
(B and C) ChIP-Seq and RNA-Seq data visualized with Integrative Genomics Viewer browser for the genomic regions surrounding the genes (B) Cyclin E (CycE), (C) delta-1-Pyrroline-5-carboxylate dehydrogenase 1 (P5CDh1), Cytochrome c proximal (Cyt-c-p), and Z band alternatively spliced PDZ-motif protein 66 (Zasp66). n = 2 samples/condition. The expression of these genes in Mef2>mCherry-RNAi muscles and Mef2>Rbf-RNAi in muscles is displayed with the reads assembled. n = 2/genotype. Read scales are (B) 0–15 in Rbf ChIP, 0–92 in Dp ChIP, and 0–44 in RNA-seq for CycE; (C) 0–29, 0–109, and 0–296 for P5CDh1; 0–33, 0–123, and 0–869 for Cyt-c-p; and 0–23, 0–85, and 0–381 for Zasp66, respectively. GroupAuto scale was used for RNA-seq.
(D) Average profile of Rbf (red) and Dp (blue) mapped reads in 50 bp plotted near the TSS in kilobase using Cis-Regulatory Element Annotation System (CEAS) for sequences repressed by Rbf (986 transcripts) and activated by Rbf (851 transcripts).
(E) De novo motif discovery MEME. Sequences repressed by Rbf (986 transcripts and 575 peaks) and activated by Rbf (851 transcripts and 404 peaks). TOMTOM matched the motif against the vertebrate database.
(F) ChIP-qPCR from pupae (96 h APF) using anti-Rbf, anti-Dp, and nonspecific antibodies (IgG and anti-Myc). Genes are P5CDh1, Cyt-c-p, and Zasp66. Fold-enrichment relative to the negative site for each ChIP sample. Mean ± SD, n = 2 replicates per antibody, two-way ANOVA, Bonferroni’s multiple comparisons test, **p < 0.01.
(G) Lysates of pupae (96 h APF) blotted against Rbf and β-tubulin antibodies. Genotypes are Mef2-GAL4/UAS-mCherry-RNAi and UAS-Dp-RNAi;Mef2-GAL4. See also Table S5.