Figure 2. Myelin outfoldings and reduced nerve conduction velocity in mice lacking oligodendroglial expression of ANLN.
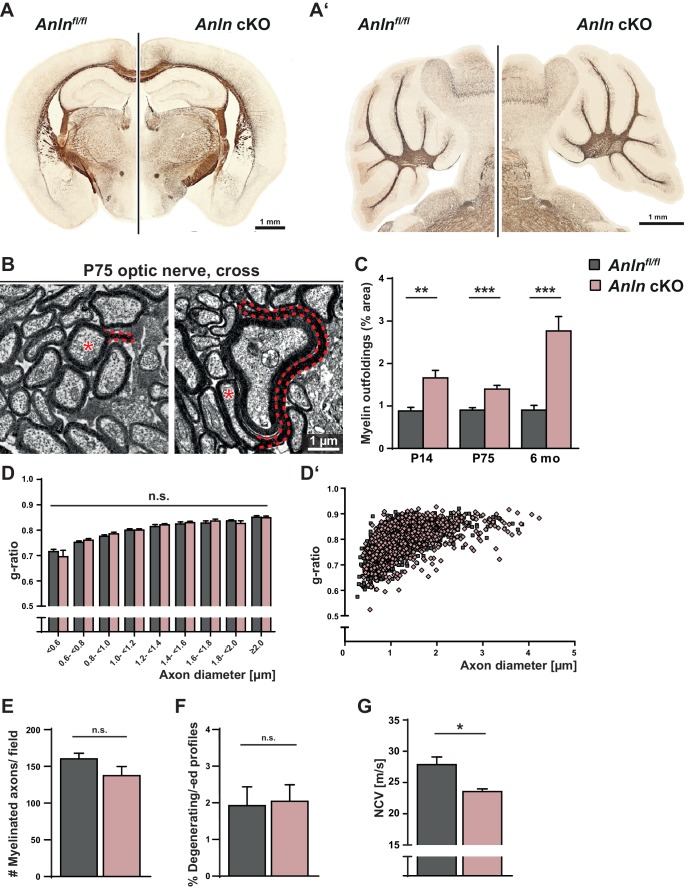
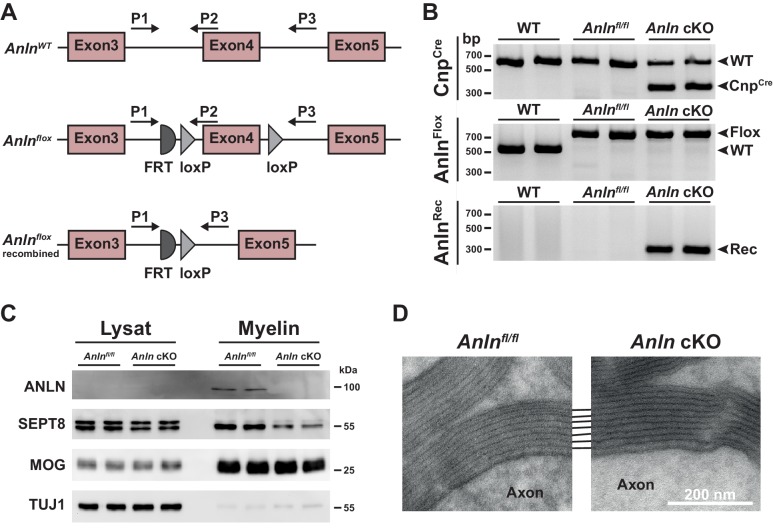
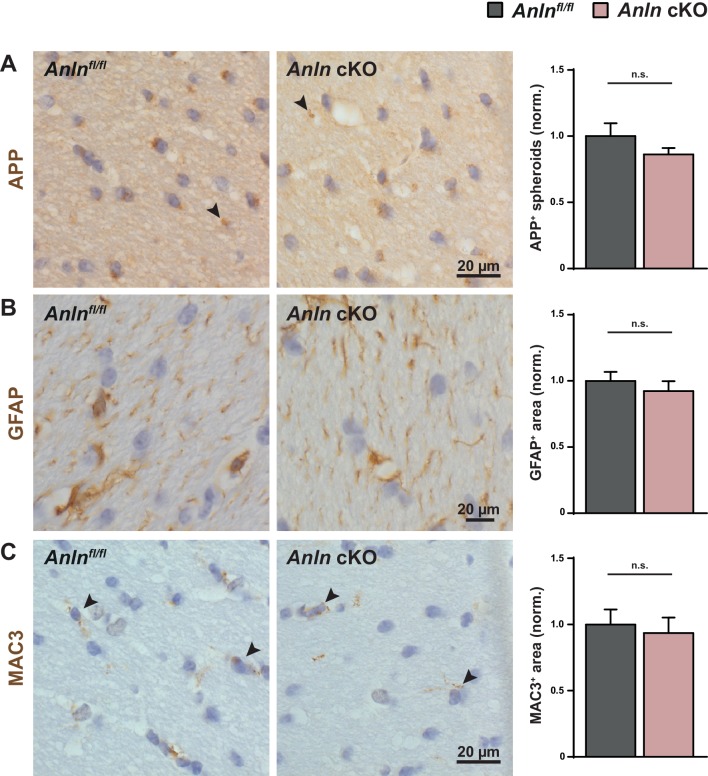
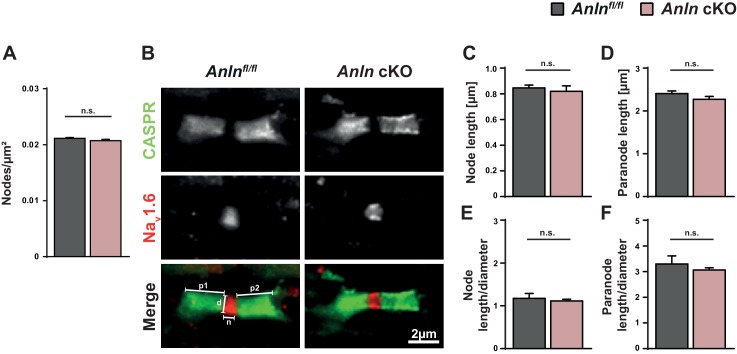
(A–A’) Silver impregnation (in brown) visualizes myelinated fiber tracts in mice lacking ANLN from myelinating cells (Anlnfl/fl;CnpCre/WT-mice; Anln cKO) and in control mice (Anlnfl/fl) at P75. (A) displays coronal brain sections; A’) shows sagittal sections through the cerebellum. Images representative of three mice per genotype. For generation and validation of Anln cKO mice see Figure 2—figure supplement 1. (B) Electron micrographs of optic nerves exemplify myelin outfoldings at P75. Stippled lines highlight myelin outfoldings; associated axons are marked with asterisks. (C) Quantitative evaluation of electron micrographs of optic nerves reveals progressive emergence of myelin outfoldings in adult Anlnfl/fl;CnpCre/WT mice (Anln cKO). Mean +/SEM. n = 4–6 mice per genotype and age; two-tailed unpaired t-test P14 p=0.0076; P75 p=0.0009; 6mo p=0.0007. (D,D‘) g-ratio analysis of electron micrographs of optic nerves at six mo indicates normal myelin sheath thickness in Anln cKO mice. Mean +/SEM. Not significant according to two-way ANOVA (p=0.9279). (E) Quantitative evaluation of electron micrographs of optic nerves at six mo reveals a normal frequency of myelinated axons in Anln cKO mice. Mean +/SEM. n = 4–5 mice per genotype; not significant (n.s.) according to two-tailed unpaired t-test (p=0.1827). (F) Quantitative evaluation of electron micrographs of optic nerves at six mo indicates that there is no increased frequency of degenerating/degenerated axons in Anln cKO mice. Mean +/SEM. n = 4–5 mice per genotype; not significant (n.s.) according to two-tailed unpaired t-test (p=0.8664). For immunohistochemical assessment of neuropathology see Figure 2—figure supplement 2. (G) Electrophysiological measurement reveals reduced nerve conduction velocity in the spinal cord of Anln cKO compared to control (Anlnfl/fl) mice at six mo. Mean +/SEM. n = 7–11 mice per genotype; two-tailed unpaired t-test (p=0.0149). For assessment of density and dimensions of the nodes of Ranvier see Figure 2—figure supplement 3.