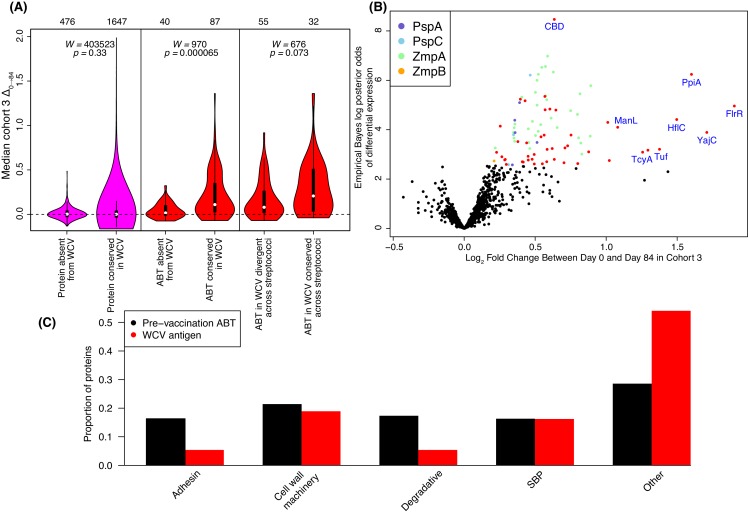
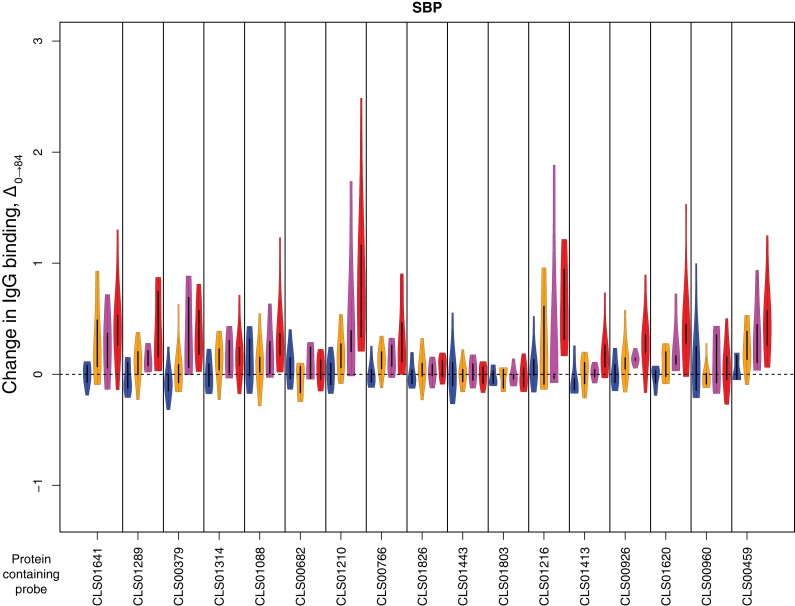
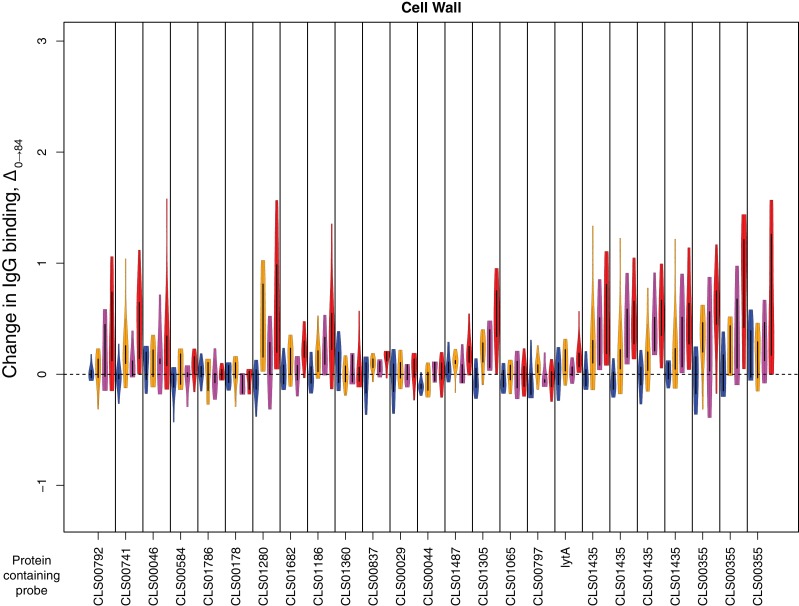
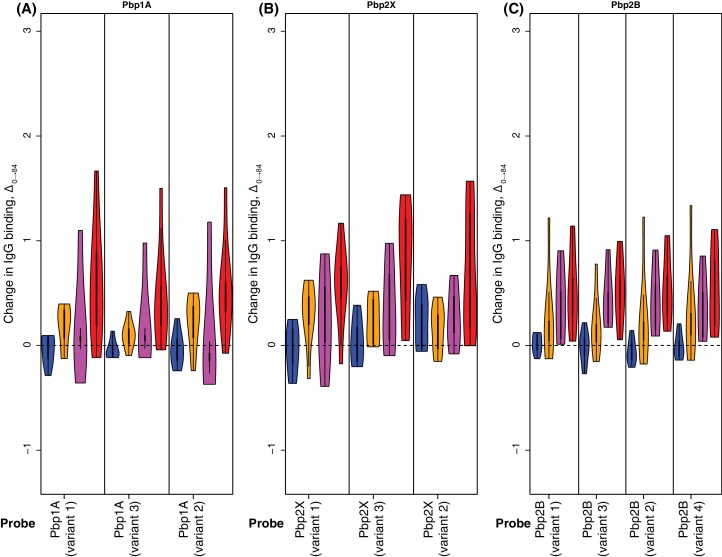
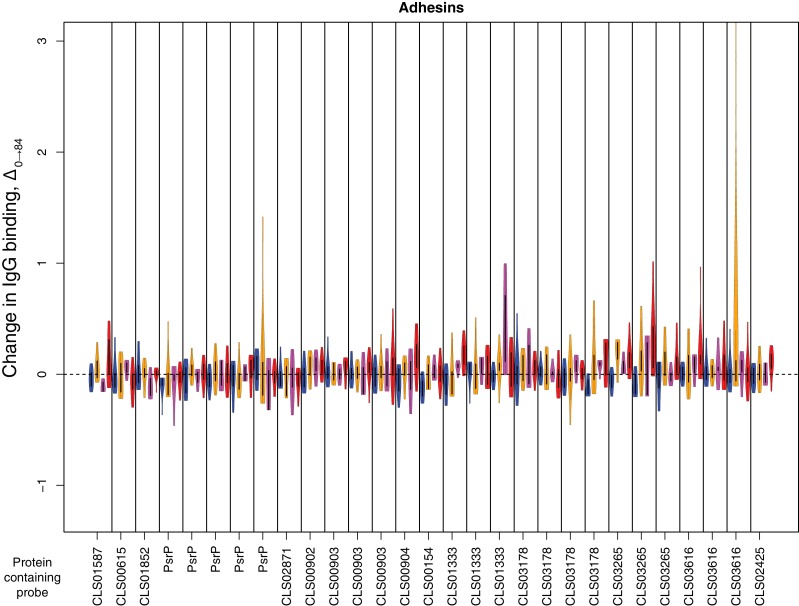
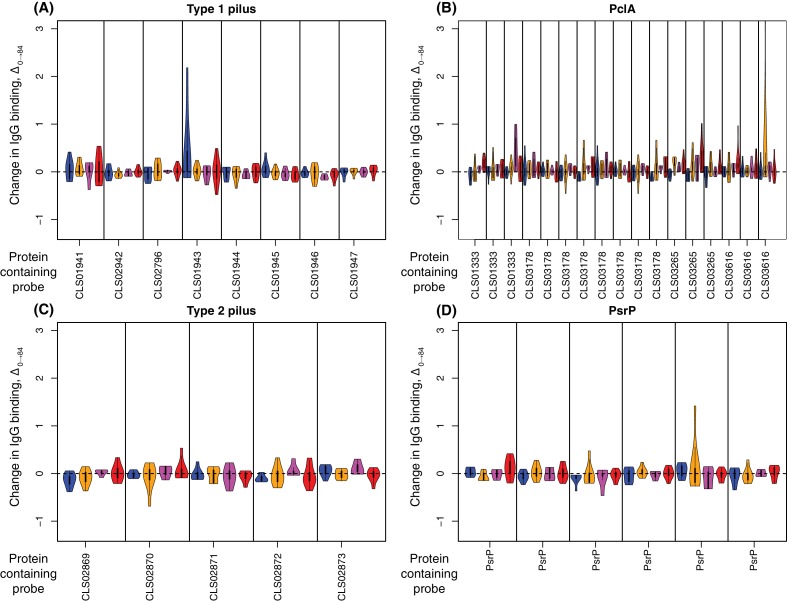
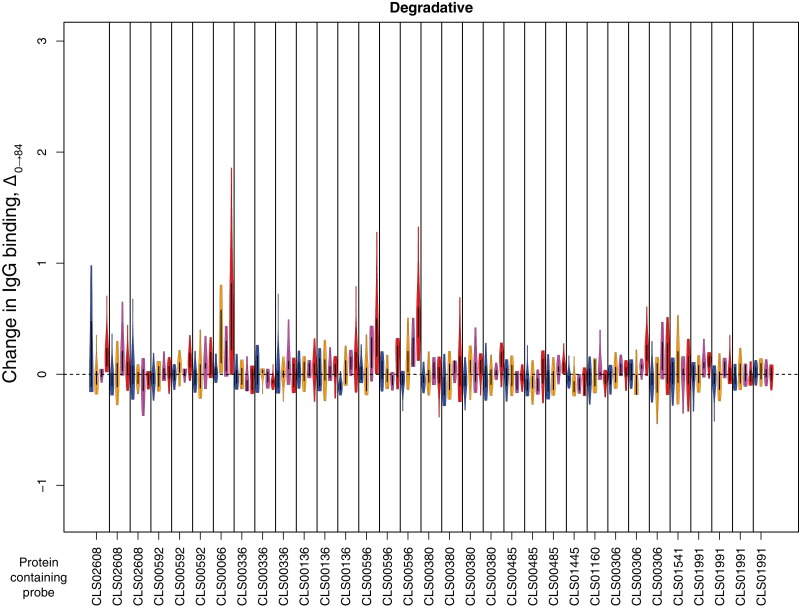
Figure 4. Identification of WCV antigens from changes in IgG binding.
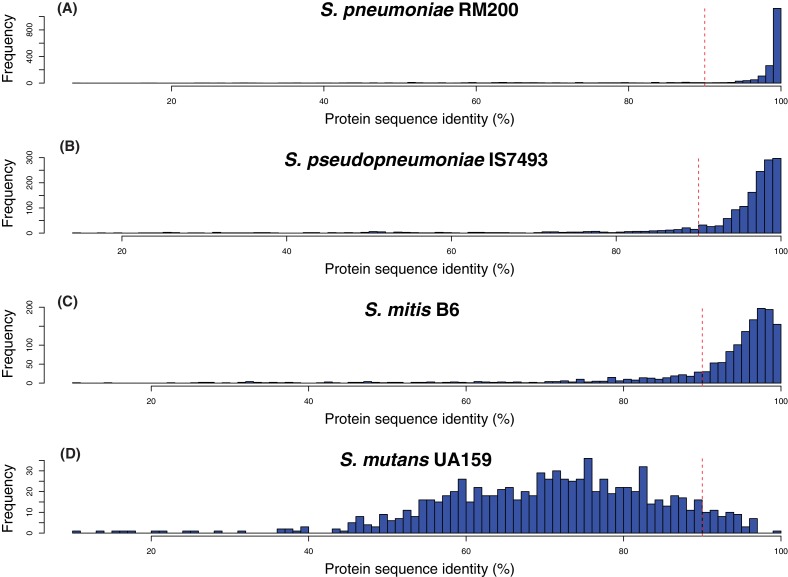
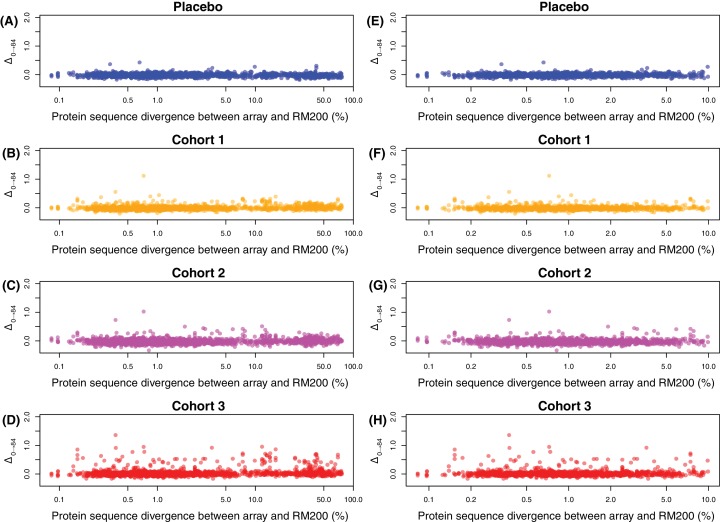
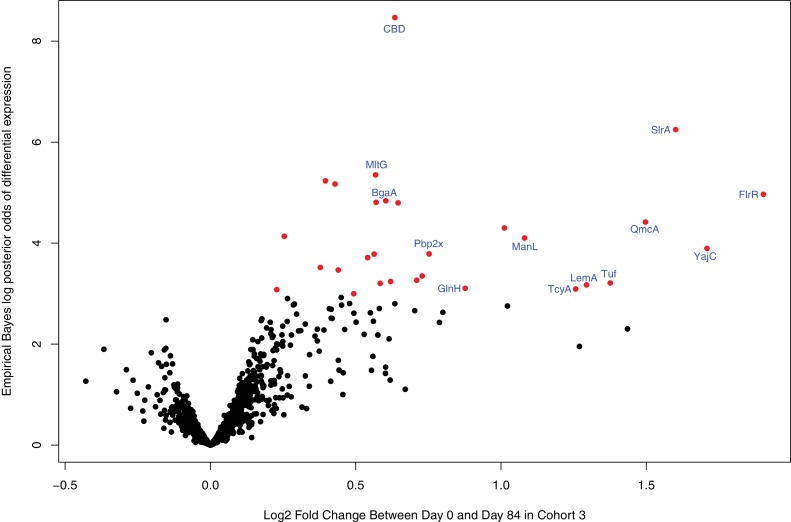
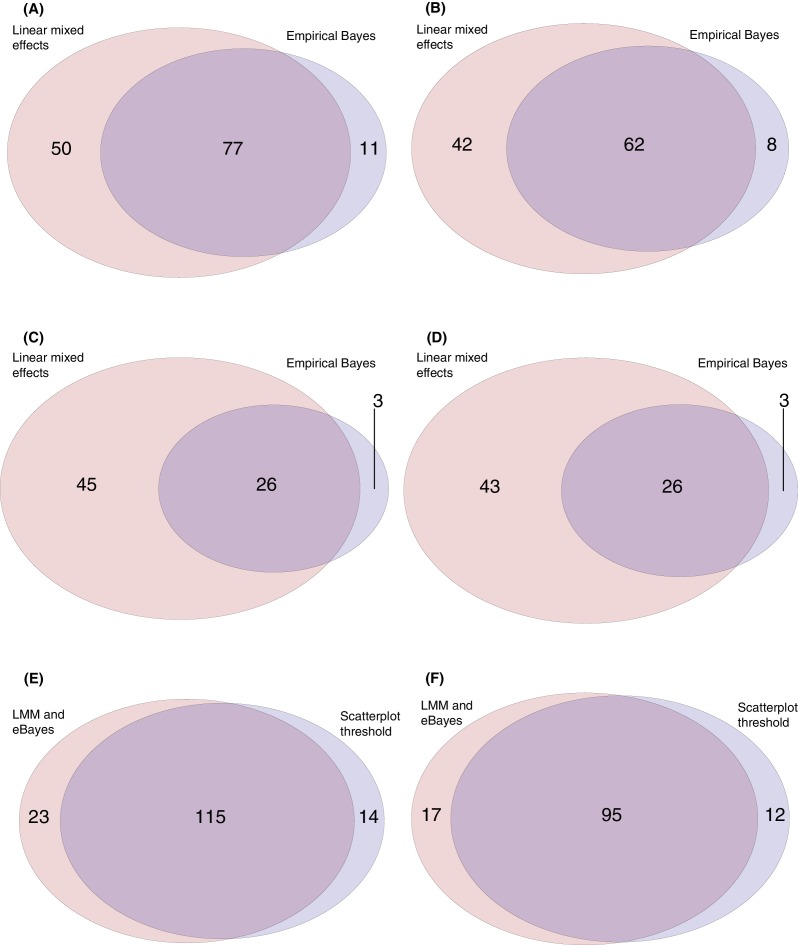
(A) Violin plots showing the median Δ0→84 within cohort three for non-DCL array probes, split by whether they represent proteins with close orthologues in the WCV strain RM200 (defined as ≥90% amino acid identity) or not. The central panel shows the same comparison, but further constraining the dataset to probes representing the proteins classified as ABTs based on high pre-vaccination IgG binding. The right panel stratifies the 87 probes corresponding to ABTs with close orthologues in RM200 by whether the protein on the array was also conserved with ≥90% sequence identity in S. mitis and S. pseudopneumoniae (Supplementary file 1). The number of probes in each category is shown at the top of the plot. The results of Wilcoxon rank sum tests (W) and significance (p) are annotated on each panel. (B) Volcano plot showing the statistical and biological significance of changes in IgG binding following WCV administration in cohort 3. The horizontal axis shows the fold change in IgG binding between the day zero and day 84 samples from cohort three on a base two logarithmic scale. The vertical axis shows the B statistic from the empirical Bayes analysis, representing the natural logarithm of the odds ratio of differential IgG binding between cohort three and the placebo group. Points corresponding to array probes with a Benjamini-Hochberg corrected p value below 0.05 are coloured red, unless they represent a variant of PspA, PspC, ZmpA, or ZmpB. (C) Functional categorisation of antigens. The distribution of ABTs, defined as eliciting high IgG in the pre-vaccination samples (Croucher et al., 2017), and the WCV antigens identified by either the eBayes or LMM analyses (Supplementary file 2) are compared across different functional categories.