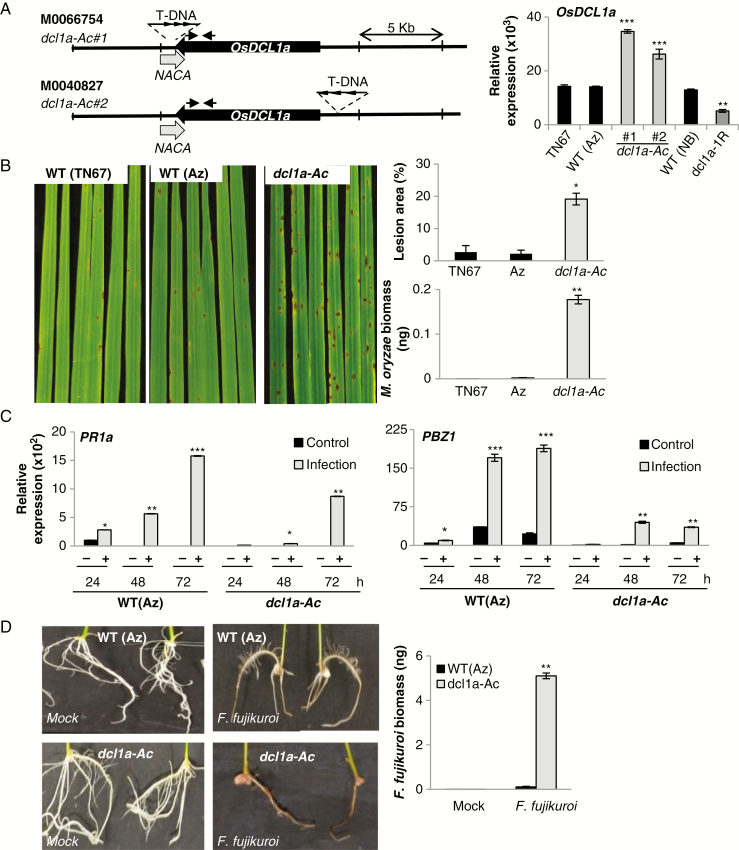
Fig. 1.
Susceptibility of OsDCL1a activation mutants to infection by the pathogens M. oryzae and F. fujikuroi. (A) Representation of the T-DNA insertion mutants from the TRIM collection (lines M0066754 and M0040827) (left panel). Black and grey arrows represent OsDCL1a (Os03g02970) and the nearby genes (Nascent polypeptide-associated complex subunit alpha, NACA; Os03g02960) pointing in the direction of transcription. Arrowheads in the T-DNA represent the CaMV35S enhancer octamers. Arrows above the OsDCL1a gene indicate the position of primers used for RT-qPCR analysis. Right panel: OsDCL1a expression in leaves of 3-week-old dcl1a-Ac and dcl1a-IR plants determined by RT-qPCR. Tainung67 (TN67) and Nipponbare (NB) are the genetic backgrounds of the TRIM mutants and dcl1a-IR plants, respectively. (B) Susceptibility of the dcl1a-Ac#1 mutant to M. oryzae infection. Three-week-old rice plants were inoculated with M. oryzae spores (1 × 105 spores mL–1). Pictures were taken at 7 days post-inoculation (dpi). Right panels show the lesion area in infected leaves (measured by Assess 2.0) and quantification of M. oryzae DNA by qPCR at 7 dpi. (C) OsPR1a and OsPBZ1 expression in wild-type (segregated azygous) and dcl1a-Ac#1 plants determined by RT-qPCR at the indicated times after inoculation with M. oryzae spores (1 × 105 spores mL–1). (D) Susceptibility of dcl1a-Ac plants to infection by F. fujikuroi. Seeds of dcl1a-Ac#1 and wild-type (segregated azygous) plants were germinated for 24 h and inoculated with F. fujikuroi spores (106 spores mL–1). Roots from F. fujikuroi-inoculated seedlings at 7 dpi are shown. Right panel: F. fujikuroi DNA in roots of wild-type and dcl1a-Ac plants was quantified by qPCR at 7 dpi. Three independent infection experiments with each fungus were performed (at least 24 plants per genotype in each experiment). Data are the mean ± s.d. (n = 3 biological replicates) (*P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001 by ANOVA).