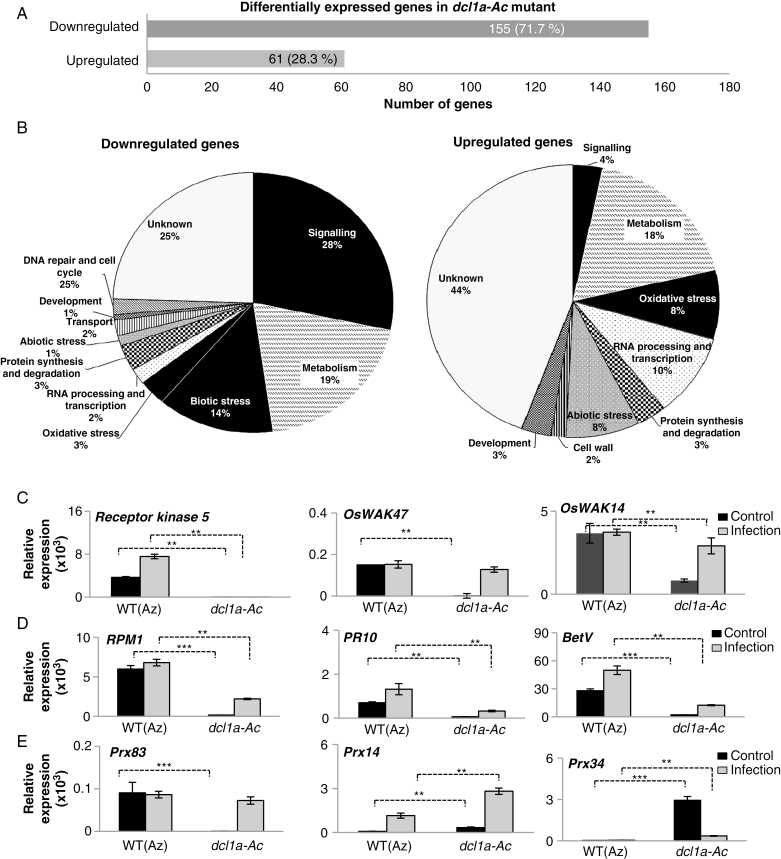
Fig. 3.
Distribution and validation of differentially expressed genes in dcl1a-Ac plants. (A) Total number of differentially expressed genes in leaves of dcl1a-Ac plants compared with wild-type plants (downregulated and upregulated genes). (B) Functional categories of downregulated and upregulated genes in leaves of dcl1a-Ac plants. (C–E) Validation and fungal responsiveness of differentially expressed genes identified by RNA-Seq. Transcript levels were determined by RT-qPCR in leaves of control (non-infected) and M. oryzae-infected plants (at 72 hpi) (black and red bars, respectively). (C) Receptor kinase 5 (Os09g37880), OsWAK47 (Os04g30260) and OsWAK14 (Os10g39680). (D) Disease resistance RPM1 (Os11g12340), PR10 (Os12g36860) and Bet V (PR10 family; Os12g36850). (E) Prx83 (Os06g32990), Prx14 (Os07g48050) and Prx34 (Os03g02939). Four biological samples (including the same RNA samples used for RNA-Seq experiments for non-inoculated plants) and two technical replicates were examined (**P ≤ 0.01; ***P ≤ 0.001 by ANOVA).