Abstract
Background and Aims
Orchidaceae is a large plant family, and its extraordinary adaptations may have guaranteed its evolutionary success. Flavonoids are a group of secondary metabolites that mediate plant acclimation to challenge environments. Chalcone synthase (CHS) catalyses the initial step in the flavonoid biosynthetic pathway. This is the first chromosome-level investigation of the CHS gene family in Phalaenopsis aphrodite and was conducted to elucidate if divergence of this gene family is associated with chromosome evolution.
Methods
Complete CHS genes were identified from our whole-genome sequencing data sets and their gene expression profiles were obtained from our transcriptomic data sets. Fluorescence in situ hybridization (FISH) was conducted to position five CHS genes to high-resolution pachytene chromosomes.
Key Results
The five Phalaenopsis CHS genes can be classified into three groups, PaCHS1, PaCHS2 and the tandemly arrayed three-gene cluster, which diverged earlier than those of the orchid genera and species. Additionally, pachytene chromosome-based FISH mapping showed that the three groups of CHS genes are localized on three distinct chromosomes. Moreover, an expression analysis of RNA sequencing revealed that the five CHS genes had highly differentiated expression patterns and its expression pattern-based clustering showed high correlations between sequence divergences and chromosomal localizations of the CHS gene family in P. aphrodite.
Conclusions
Based on their phylogenetic relationships, expression clustering analysis and chromosomal distributions of the five paralogous PaCHS genes, we proposed that expansion of this gene family in P. aphrodite occurred through segmental duplications, followed by tandem duplications. These findings provide information for further studies of CHS functions and regulations, and shed light on the divergence of an important gene family in orchids.
Keywords: Chalcone synthase, gene family, gene duplication, expression profile, chromosome evolution, fluorescence in situ hybridization
INTRODUCTION
Orchidaceae is a large plant family among the angiosperms, with >25 000 species that belong to >800 genera (Dressler, 1993). Orchids show high levels of variation in their floral morphologies, which make them, especially the Phalaenopsis cultivars, popular ornamentals in global horticultural markets. In addition, orchids are epiphytic, terrestrial or even lithophytic, and are renowned for their extraordinary acclimations. The evolutionary success of Orchidaceae may result from their diversified reproductive and ecological strategies (Givnish et al., 2015).
Flavonoids are a group of plant secondary metabolites. They play numerous important roles in plants, such as flower pigmentation (anthocyanins), legume–rhizobial interactions (isoflavones), protection against UV radiation (flavonols), pathogen defence (isoflavonoids) and pollen fertility (Ferreyra et al., 2012). Thus, they greatly enhance plant tolerance and adaptive ability. The flavonoid biosynthetic pathway includes the phenylpropanoid and flavonoid pathways.
Chalcone synthase (CHS) is a key enzyme in the phenylpropanoid biosynthetic pathway and is ubiquitous in plants. It functions in the initial step of the phenylpropanoid pathway and condenses three malonyl-CoA molecules with one 4-coumaroyl-CoA molecule to generate a naringenin chalcone, which is the precursor of various flavonoids. Owing to their importance in flavonoid production, CHS genes have been well studied in plant species such as Petunia hybrida (Koes et al., 1989), Zea mays (Franken et al., 1991; Han et al., 2016), Ipomoea purpura (Durbin et al., 1995, 2000), Gerbera hybrida (Helariutta et al., 1996; Deng et al., 2014), Arabidopsis thaliana (Saslowsky et al., 2000), Dendranthema species (Yang et al., 2002), Viola species (van den Hof et al., 2008), Physcomitrella patens (Koduri et al., 2010), 12 species belonging to the Rosid clade (Zavala and Opazo, 2015) and Oryza sativa (Hu et al., 2017).
To date, most of the examined plant gene families originated through gene duplication. Gene copies can arise via several mechanisms (Panchy et al., 2016). Whole-genome duplication or the duplication of an entire chromosome results in the most dramatic form of gene duplication, but there are also small-scale sub-genomic duplication events, such as tandem duplication, segmental duplication, transposon-mediated duplication and retroduplication. The duplicated genes aid plant acclimation in the following three main ways: (1) they increase the production of beneficial products; (2) they may generate newly functional genes; and (3) they may specialize in spatial and developmental expression.
Gene duplication followed by divergence is a conspicuous feature of plant gene families, especially in genes related to plant secondary metabolism (Ober, 2005). For example, shifts in substrate specificity and catalytic reaction were observed in GhCHS2 of G. hybrida (Helariutta et al., 1996). Additionally, expression patterns of duplicate copies of CHS genes resulted in extensive differentiations in I. purpura (Durbin et al., 2000), Vitis vinifera (Goto-Yamamoto et al., 2002), O. sativa (Hu et al., 2017), Z. mays (Han et al., 2016) and P. patens (Koduri et al., 2010). Those findings demonstrated that the duplicate CHS genes play specialized functional roles over the course of evolution. The structures of CHS genes examined to date have been conserved, and exhibited two exons and one intron, except for the CHS genes of Antirrhinum majus (Sommer and Saedler, 1986), which had two introns, and a few CHS genes of Z. mays and P. patens, which were intronless (Koduri et al., 2010; Han et al., 2016). Recent advances in high-throughput sequencing techniques and the construction of physical maps have made conducting genome-wide surveys easier. Thus, genome-wide identification, characterization, expression and distribution analyses of the CHS gene family have been conducted in Z. mays and O. sativa (Han et al., 2016; Hu et al., 2017), and the evolution of this gene family should be further investigated in other species.
Studies of the CHS gene family in orchids have been limited. Three CHS genes were cloned from the flower of the orchid Bromheadia finlaysoniana (Liew et al., 1998). OCHS3 was highly expressed in young leaves, which were flushed with anthocyanin, and was detected at much lower levels in faintly coloured flowers. Pitakdantham et al. (2010) also isolated a CHS gene from Dendrobium Sonia Earsakul, and it was highly expressed in young flowers. In Phalaenopsis, three CHS genes were isolated, and their expression patterns in floral tissues at different developmental stages were diverse (Han et al., 2006). Of the three CHS genes, PhCHS5 was most highly expressed and was the sole CHS gene responsible for pigment accumulation. Additionally, the orchid CHS genes were further grouped into two clades, Orchid CHS1 and Orchid CHS2 groups, based on amino acid sequences. Chomicki et al. (2015) demonstrated that two CHS genes under UV-B induction were expressed in orchid root tips for effective protection of the photosynthetic cortex. However, comprehensive investigations of the CHS genes in orchid species that analyse their gene duplications, distributions, evolutionary processes and functional diversifications in multiple tissues/organs at different developmental stages are scarce. In addition, numerous transcriptomic databases are available (Su et al., 2013; Tsai et al., 2013; Cai et al., 2015; Chao et al., 2017); we are currently conducting whole-genome sequencing, assembling and annotation to facilitate an overall survey of this gene family.
Here, a comprehensive study of the CHS gene family in Phalaenopsis aphrodite was conducted. The CHS genes were identified by searching transcriptomic data sets, and their phylogenetic relationships with CHS genes in other plants were revealed. Furthermore, the expression profiles in various tissues/organs and developmental stages, and chromosomal localizations were analysed. These results provide a foundation for further study of the CHS gene family in orchids and an example of gene localization and functional diversification in Orchidaceae.
MATERIALS AND METHODS
Identification of CHS genes in Phalaenopsis species and phylogenetic analysis
The CHS genes of P. aphrodite were identified from our orchid transcriptomics database, Orchidstra 2.0 (Chao et al., 2017). The identities of amino acid and nucleotide sequences of the coding regions and sequence alignments were analysed using Vector NTI (Li and Moriyama, 2004). A phenetic tree was constructed based on the amino acid sequences of 60 CHSs from different plant taxa (Supplementary Data Table S1). The CHS sequences of P. modesta and P. equestris were downloaded from Orchidstra 2.0 (Chao et al., 2017) and OrchidBase 2.0 (Tsai et al., 2013), respectively, and the CHS sequences of the other plant species were downloaded from NCBI’s GenBank. These amino acid sequences were aligned using default the setting in ClustalW (Thompson et al., 1994) implemented in MEGA7.0 (Kumar et al., 2016). The P. patens CHS gene was used as the outgroup. Evolutionary history was inferred using the Neighbor–Joining (NJ) (Saitou and Nei, 1987) method. Evolutionary distances were computed using the Poisson correction method (Zuckerkandl and Pauling, 1965), and node support was assessed by bootstrap test (Felsenstein, 1985) with 1000 resampling replicates using MEGA7.0 (Kumar et al., 2016).
Cloning and labelling of DNA probes for fluorescence in situ hybridization (FISH) mapping
The cDNA sequences of CHS genes in P. aphrodite derived from the Orchidstra 2.0 database were first used as queries in a BLAST algorithm-based search of the assembled genomic shotgun sequences of P. aphrodite. Primers were designed using Primer3 to amplify DNA fragments that included or were close to the CHS coding regions from the genomic DNA of P. aphrodite (Supplementary Data Table S2). The CHS-containing DNA fragments were amplified by PCR, which was performed using 10 ng of genomic DNA, 1× PCR buffer, 0.4 mm of each dNTP, 0.3 μm each of forward and reverse primer, and 1.0 U of KOD FX Neo DNA polymerase (TOYOBO, Osaka, Japan) in a total reaction volume of 50 μL. The PCR step-down cycling conditions were as follows: 94 °C for 2 min; five cycles of 98 °C for 10 s and 70 °C for 3.5 min; five cycles of 98 °C for 10 s and 68 °C for 3.5 min; five cycles of 98 °C for 10 s and 65 °C for 3.5 min; 20 cycles of 98 °C for 10 s and 60 °C for 3.5 min; 68 °C for 7 min; and then 25° C for 10 s. The PCR products were then cloned into the pZeroback vector (TIANGEN Biotech, Beijing, China) and transformed into Escherichia coli DH5α.
Plasmid DNA was extracted using a Plasmid Miniprep Purification Kit (GMbiolab, Taichung, Taiwan), and 1.8 μg of plasmid DNA was labelled with either biotin-dUTP or digoxigenin-dUTP using standard nick translation following the manufacturer’s protocol (Roche, Basel, Switzerland). Three plasmid DNAs with a total DNA insert size of approx. 10 kb were mixed together and used as a probe for pachytene chromosome-based FISH mapping (Supplementary Data Table S2).
Preparation of pachytene chromosomes and FISH mapping
Flower buds of P. aphrodite (2n = 2x = 38) with a size range of 8.50–9.20 mm were collected to obtain developing pollinia with chromosomes at the meiotic pachytene stage. Pachytene chromosomal spreads were prepared following the modified drop method, as previously described (Kuo et al., 2016). Meiotic pachytene spreads with little to no cytoplasm and good pachytene chromosome spreading were selected and stored at 4 °C for later use.
The FISH was carried out as previously described, with some modifications (Zhong et al., 1996). The selected slides were incubated at 37 °C overnight or at 65 °C for 30 min to air-dry the chromosomal spreads before FISH. The pre-hybridization treatment included 5 μg mL–1 pepsin for 20 min and freshly prepared formaldehyde buffer [1× phosphate-buffered saline (PBS), 50 mm MgCl2 and 1 % formaldehyde] for 10 min. Slides were then dehydrated through an ethanol series (70, 90 and 100 %). The hybridization mixture (10 % dextran sulphate sodium, 50 % formamide, 2× SSC, 0.25 % SDS and 100–200 ng of probe DNA) was boiled for 10 min, placed on ice for 5 min and added onto the slides. The slides were treated at 80 °C for 2.5 min on a hot plate and incubated in a humid chamber at 37 °C for 12–16 h. Digoxigenin-labelled probes were detected and amplified with sheep fluorescein isothiocyanate-conjugated anti-digoxigenin antibody (Roche) and anti-sheep–fluorescein isothiocyanate (VECTOR Laboratories, Burlingame, CA, USA), respectively, whereas biotin-labelled probes were detected and amplified with Avidin Texas Red (VECTOR Laboratories) and biotinylated anti-avidin D (VECTOR Laboratories), respectively. Finally, slides were dehydrated through an ethanol series (70, 90 and 100 %) and dried at 37 °C for 20 min. Chromosomes were counterstained with 1.5 μg mL–1 4’,6-diamidino-2-phenylindole in mounting medium (VECTOR Laboratories). Images were captured using a Nikon DS Ri1 CCD camera attached to a Nikon ECLIPSE 80i microscope (Nikon, Tokyo, Japan). Images were adjusted using NIS-Elements D3.2 and Adobe Photoshop CS3. Chromosome straightening was carried out using ImageJ (https://imagej.nih.gov.ij/).
Digital expression analysis
Orchidstra 2.0 (Chao et al., 2017) is a publicly available transcriptomics resource for orchid species. Normalized transcripts per million (TPM) values derived from RNA sequencing (RNA-seq) were searched and downloaded from Orchidstra 2.0. Nine TPM values for each of the five P. aphrodite CHS genes examined in different tissues/organs or at different developmental stages were obtained. The downloaded TPM values were pre-processed into the logarithm of TPM to base 2 and converted into a heatmap by ClustVis (Metsalu and Vilo, 2015).
RESULTS
Identification of CHS genes in Phalaenopsis species
A great number of orchid sequences have become publicly available in databases, and they are valuable resources for orchid gene identification. In this study, we searched for CHS genes of P. aphrodite using an orchid transcriptomic database, Orchidstra 2.0 (Chao et al., 2017). Five annotated CHS genes of P. aphrodite were identified and named PaCHS1 to PaCHS5 (Table 1). The length of their coding regions ranged from 388 (PaCHS2) to 395 (PaCHS1) amino acids, and these sequences were aligned (Supplementary Data Fig. S1). The defining amino acids of the CHS family and reported active site residues were all conserved in the five CHSs (Ferrer et al., 1999). The identities of amino acid sequences ranged from 60 to 96 % (Table 1), and the identities of nucleotides in the coding regions were only slightly different from those of the amino acid sequences. The identities between the PaCHS1 sequence and the other four CHS sequences were relatively low, and ranged from 60 to 66 %. The highest protein sequence identity was between PaCHS4 and PaCHS5 (96 %). In addition, the sequence identities among PaCHS3, PaCHS4 and PaCHS5 were all higher than 87 %, and these sequences grouped into a cluster (Supplementary Data Fig. S2). Thus, based on the protein sequences, the five CHSs of P. aphrodite were classified into three groups, which consisted of PaCHS1, PaCHS2 and another three CHSs showing high sequence identities. In addition, the five CHS genes were all composed of two exons and one intron with similar gene structures and sizes.
Table 1.
Sequence identities of the five chalcone synthase (CHS) genes in P. aphrodite
| Sequence ID | Gene | Length of coding regions | Identity of amino acid and nucleotide sequences (%) | ||||
|---|---|---|---|---|---|---|---|
| PaCHS1 | PaCHS2 | PaCHS3 | PaCHS4 | PaCHS5 | |||
| PATC124207 | PaCHS1 | 395 | – | 60 | 63 | 66 | 64 |
| PATC125513 | PaCHS2 | 388 | 64 | – | 70 | 71 | 70 |
| PATC159204 | PaCHS3 | 391 | 63 | 69 | – | 92 | 91 |
| PATC125905 | PaCHS4 | 391 | 64 | 69 | 88 | – | 96 |
| PATC124475 | PaCHS5 | 393 | 63 | 68 | 87 | 93 | – |
The italic values are for nucleotide sequences.
Chromosomal localizations of CHS genes in P. aphrodite
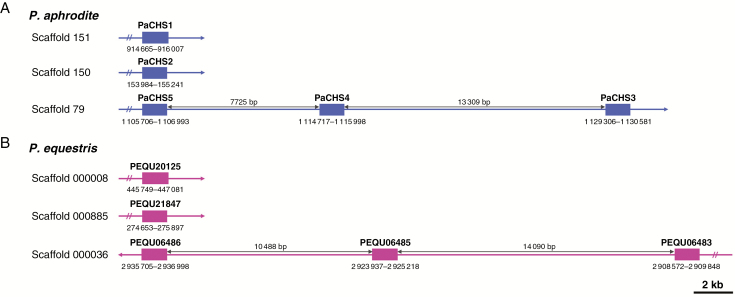
During evolution, tandem and segmental duplications are considered the main driving forces that expand gene families. Here, the distribution of the five CHS genes in chromosomes was investigated to determine the possible occurrence of duplication events that affected expansion of CHS genes in P. aphrodite. The coding sequences of the CHS genes were first mapped onto the assembled genome sequences of P. aphrodite. PaCHS3, PaCHS4 and PaCHS5 were tandemly arrayed on the same scaffold, scaffold 79 (Fig. 1A). PaCHS1 and PaCHS2 were located on scaffolds 151 and 150, respectively. However, neither genetic linkage nor physical maps have been established for Phalaenopsis species. Thus, FISH mapping was the technique chosen to investigate localizations of the CHS genes to P. aphrodite chromosomes.
Fig. 1.
Physical positions of the CHS genes on the Phalaenopsis assembled genome sequences. The positions, scaffold numbers and intervals between the three tandemly arrayed CHS genes in (A) P. aphrodite and (B) P. equestris are indicated.
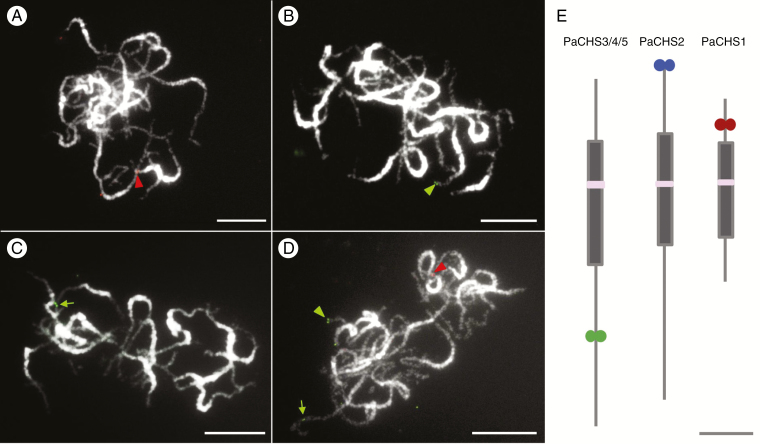
Meiotic pachytene chromosomes, instead of mitotic metaphase chromosomes, were applied to enhance FISH mapping resolution. DNA fragments including the CHS genes and nearby fragments were amplified, cloned and then used as scaffold-specific probes to detect their precise locations on pachytene chromosomes. The primers used to amplify the DNA fragments are listed in Supplementary Data Table S2. For all three probes, PaCHS1, PaCHS2 and PaCHS3/4/5, only one distinct signal was detected on one of the 19 P. aphrodite chromosomes (Fig. 2). Probe PaCHS1 was mapped to the euchromatic region of the short arm of a pachytene chromosome (Fig. 2A), whereas probe PaCHS2 was located at the end of the short arm of a chromosome (Fig. 2B). Probe PaCHS3/4/5, which represented three tandemly arrayed CHS genes, was detected within the euchromatic region of the long arm of a chromosome (Fig. 2C). Furthermore, a pool of the three probes showed that they were located on three different P. aphrodite chromosomes (Fig. 2D). The ideograms of the three CHS gene-containing chromosomes are illustrated in Fig. 2E.
Fig. 2.
FISH mapping of CHS genes on pachytene chromosomes of P. aphrodite. PaCHS1 (A, red arrowhead), PaCHS2 (B, green arrowhead) and PaCHS3/4/5 (C, green arrow) only showed distinct signals on a P. aphrodite pachytene chromosome. (D) A pool of the three CHS probes in (A–C). Chromosomes were stained with 4’,6-diamidino-2-phenylindole, and images were converted to black and white. Scale bar = 10 μm. (E) Ideogram of the three CHS-localized pachytene chromosomes. Grey and pink boxes indicate the heterochromatic region and centromere, respectively. The green and red circles represent the chromosomal localizations of the CHS probes. Scale bar in (E) represents 5 μm.
Distinct expression profiles of the CHS genes in P. aphrodite
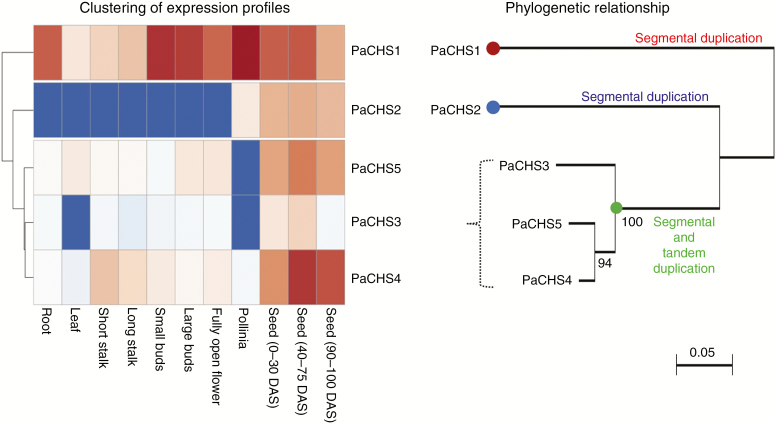
Because the duplicated CHS genes showed divergent sequences and were located on different chromosomes, they may generate sub- or neo-functions. The expression patterns of genes within tissues or organs are usually correlated with their biological functions. The expression profiles of the five P. aphrodite CHS genes investigated by RNA sequencing (RNA-seq) were obtained from Orchidstra 2.0 (Chao et al., 2017). The normalized expression data were converted to heatmaps for better visualization and their expression profiles in diverse tissues/organs at different developmental stages were also clustered (Fig. 3). PaCHS1 showed constitutive elevated expression levels among organs and developmental stages. The highest and lowest PaCHS1 expression levels were in pollinia and leaves, respectively, and its expression level decreased from small buds (1.3 cm) to fully open flowers, which resembled previously reported PhCHS5 expression in a Phalaenopsis hybrid (Han et al., 2006). PaCHS2 had the lowest expression level of the five CHS genes, and it was only expressed in pollinia and seeds. Alternatively, the expression levels of PaCHS3, PaCHS4 and PaCHS5 were relatively lower than that of PaCHS1 and were expressed mainly in seeds. PaCHS3 and PaCHS5 expression was not detected in pollinia. Moreover, the expression patterns of PaCHS3, PaCHS4 and PaCHS5 were similar and were distinct from those of PaCHS1 and PaCHS2. Thus, the expression levels of the five CHS genes were highly differentiated and showed distinct expression patterns in P. aphrodite. Interestingly, we found that clustering based on the expression patterns of the five CHS genes was correlated with their sequence divergences and chromosomal distributions (Fig. 3).
Fig. 3.
Clustering analysis based on expression profiles of the five P. aphrodite CHS genes and its association with sequence divergence. The heatmaps were generated from the normalized transcripts per kilobase million values derived from the RNA-seq analysis. Each column represents different tissues/organs at different developmental stages. DAS, days after sowing; long stalk, 1.5–3.0 cm; short stalk, <1.0 cm. We propose that the multiple copies of PaCHS genes, PaCHS1, PaCHS2 and PaCHS3, were generated by segmental duplication followed by the event whereby PaCHS3/4/5 were arranged with additional tandem duplication.
Phenetic analysis
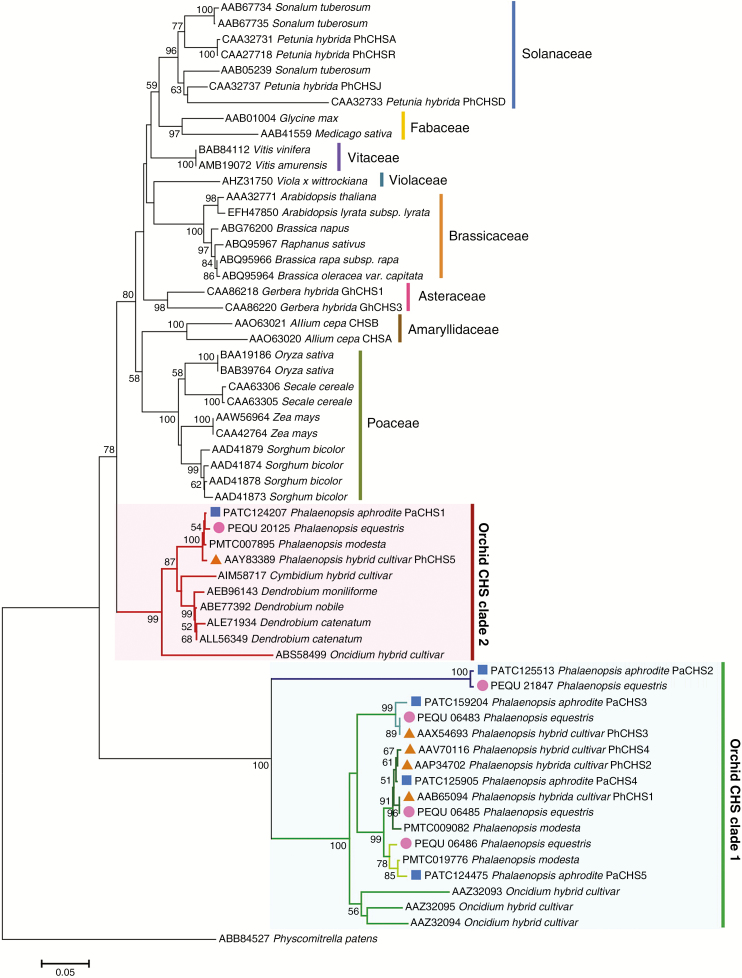
To verify the CHS gene lineage of P. aphrodite and other plant species, an NJ tree was constructed based on amino acid sequences. The detailed sequence information is listed in Supplementary Data Table S1. All orchid CHSs were classified into two major clades, Orchid clades 1 and 2 (Fig. 4), which was similar to previous findings (Chomicki et al., 2015). Of the five P. aphrodite CHSs, only PaCHS1 was included in clade 2, and it was closely related to AAY83389 (PhCHS5 of a Phalaenopsis hybrid cultivar), which is associated with anthocyanin accumulation and photosynthetic root cortex protection in Phalaenopsis cultivars (Han et al., 2006). In clade 1, PaCHS2 of P. aphrodite only clustered with PEQU_21847 of P. equestris and was distant from the other orchid CHSs in this clade. Its long branch length indicated a higher rate of sequence changes. Alternatively, the other three P. aphrodite CHSs, PaCHS3, PaCHS4 and PaCHS5, clustered in the Phalaenopsis-specific subclade. None of the orchid CHSs in the phenetic tree formed genus- or species-specific subclades, which indicates that the divergence of the orchid CHS genes might be earlier than that of the orchid genera or species. There are five homologous CHS genes in current studied Phalaenopsis species or cultivars (Fig. 4; Supplementary Data Fig. S3). In our analysis, we found five paralogous CHS genes and each had a corresponding orthologue between P. aphrodite and P. equestris, but that relationship was not found with the Phalaenopsis hybrid cultivar. There were two missing paralogues that corresponded to PaCHS2 and PaCHS5 in the Phalaenopsis hybrid cultivar, whereas three orthologues in that cultivar (PhCHS1, PhCHS2 and PhCHS4) clustered with P. aphrodite PaCHS4. These results indicate that the cultivar has a mixed and complicated set of CHS genes donated by the parental species resulting from its hybrid origin.
Fig. 4.
Neighbor–Joining tree depicting relationships among CHSs of orchids and other plant species. The Neighbor–Joining tree was constructed based on CHS amino acid sequences of P. aphrodite (blue squares), P. equestris (pink circles) and P. hybrid cultivar (orange triangles).
DISCUSSION
Flavonoids are important in plant development and acclimation. The CHS genes function in the initial step of the flavonoid biosynthetic pathway, and they are plant polyketide synthases (PKSs), which are involved in the biosynthesis of various secondary metabolites. This was the first integrated gene–chromosome–genome study of CHS genes in Orchidaceae, and five complete CHS genes were identified from the P. aphrodite transcriptomic data sets. In P. aphrodite, the CHS genes formed a relatively small gene family compared with other plant species; for example, there are three CHS genes in Viola species (van den Hof et al., 2008), six in Ipomoea species (Durbin et al., 2000), seven in Sorghum bicolor (Lo et al., 2002), eight in both Petunia hybrida (Koes et al., 1989) and Pisum sativum (Ito et al., 1997), and up to 14 in Z. mays (Han et al., 2016). However, only one complete CHS gene has been identified in A. thaliana (Saslowsky et al., 2000).
Gene duplication is considered the mechanism that leads to expansion of gene families, and it is a prevailing feature in plant genomes. Generally, a high number of gene copies in a gene family is maintained through large-scale segmental duplication or small-scale tandem duplication during evolution. In P. aphrodite, PaCHS3, PaCHS4 and PaCHS5 formed a tandemly arrayed gene cluster, and the intervals between the three CHS genes were approx.13.3 kb (PaCHS3 and PaCHS4) and 7.7 kb (PaCHS4 and PaCHS5) (Fig. 1A). This arrangement was also observed in a closely related orchid, P. equestris, in which the three CHS genes were all positioned on scaffold 000036 (Fig. 1B). Furthermore, the three P. aphrodite CHS genes were clustered in a distinct subclade of clade 1 with the three CHSs of P. equestris (Fig. 4; Supplementary Data Fig. S3). Thus, the tandem array of three CHS genes was probably present in a common ancestor before speciation within Phalaenopsis. Tandem gene duplications represent a substantial proportion of all plant genes (Jander and Barth, 2007), including 17 % in A. thaliana (Arabidopsis Genome Initiative, 2000), 14 % in O. sativa (International Rice Genome Sequencing Project, 2005), 16 % in Populustrichocarpa (Tuskan et al., 2006) and 35% in Z. mays (Messing et al., 2004). In addition, most tandem duplications, including 75 and 79 % in A. thaliana and O. sativa, respectively, contain only two genes, and tandem gene arrays with more than three gene members are rare (Rizzon et al., 2006). However, three tandemly arrayed CHS genes were found in our analysis, which indicated that tandem duplications most probably play a crucial role in expansion of Phalaenopsis CHS genes. The orchid genome harbours many remnants of one or more large-scale duplication events, but only 3.51 % of the P. equestris genome showed collinearity, which demonstrated a high degree of gene reshuffling after duplications (Cai et al., 2015). Furthermore, FISH mapping showed that PaCHS1, PaCHS2 and the gene cluster were located on three different chromosomes (Fig. 2). Thus, a large-scale genome duplication followed by reshuffling and sequence diversification might also have resulted in the duplications of Phalaenopsis CHS genes prior to the mentioned tandem duplication event. In Z. mays, no tandemly duplicated CHS genes have been found, and segmental duplication was the dominant contributor to expansion of the Z. mays CHS gene family (Han et al., 2016). In contrast, no genes emerged through segmental duplications of the O. sativa PKS gene family, and expansion of the O. sativa PKS gene family was most probably caused by tandem duplications (Hu et al., 2017). Consequently, the mechanisms underlying CHS gene duplications differed among plant species.
Chomicki et al. (2015) classified orchid CHS genes into two clades, Orchid CHS clade 1 and Orchid CHS clade 2, and these CHS genes identified from Phalaenopsis hybrid cultivars were included in our phylogenetic analysis. The NJ tree revealed that PaCHS1 clustered together with PhCHS5 (AAY83389), which belonged to Orchid CHS clade 2, whereas PhCHS1 (AAB65094), PhCHS2 (AAP34702), PhCHS3 (AAX54693) and PhCHS4 (AAV70116), members of Orchid CHS clade 2, clustered with the other four P. aphrodite CHS genes. Thus, based on the classification, PaCHS1 should belong to Orchid CHS clade 2 and the other four P. aphrodite CHSs should belong to Orchid CHS clade 2. However, the amino acid identities of PaCHS2 with the other four CHSs only ranged from 60 % (PaCHS1) to 70 % (PaCHS3, PaCHS4 and PaCHS5), and PaCHS2 only formed a subclade with a P. equestris CHS in clade 1 (Fig. 4; Supplementary Data Fig. S3). Additionally, PaCHS2 was located on a distinct chromosome (Fig. 2), and was solely expressed in pollinia and seeds; no expression was detected in the other examined tissues or organs, which was a unique expression pattern. Therefore, based on the sequence identity, phylogenetic relationships, chromosomal localization and gene expression pattern, we strongly speculate that the Phalaenopsis CHS genes diverged into three groups instead of two.
Based on the phylogenetic relationships, PaCHS1 was the only CHS gene in P. aphrodite allocated to Orchid CHS clade 2. PaCHS1 was closely related to PhCHS5 (AAY83389), which is associated with anthocyanin accumulation in the petals of Phalaenopsis cultivars (Han et al., 2006). Additionally, the PhCHS5 expression level decreased from small flower buds to fully open flowers, and this expression pattern was consistent with that of PaCHS1 observed in RNA-seq data (Fig. 3). The highly similar expression patterns between PaCHS1 and PhCHS5 may indicate analogous gene functions. However, the number and expression patterns of CHS genes may not be the only factors that determine pigment accumulation and patterning in plants. No correlation has been found between flower colour and CHS genes in Dendranthema (Yang et al., 2002), but gene inactivation and loss in the anthocyanin pathway of Iochroma (Solanaceae) is involved in flower pigmentation (Smith and Rausher, 2011; Smith et al., 2013). Additionally, the combined expression of three MYB transcription factors that regulate pigmentation patterning was reported in Phalaenopsis cultivars (Hsu et al., 2015). Therefore, even though PaCHS1 is highly expressed in the stalks, flower buds and fully open flowers of P. aphrodite, the flowers are more or less white, with only a minor yellow pigmentation on the lips.
In addition to sequence divergences, spatial and developmental expression patterns of duplicate genes can differ. Distinct gene expression patterns may reflect different physical and chemical features, functions and regulations. The recently launched transcriptomic data sets provide valuable resources for analysing orchid functional genes in a genome scale. The normalized TPM values derived from P. aphrodite RNA-seq provided comparable data for analysing gene expression levels in various plant tissues or organs at different stages. The P. aphrodite CHS genes, with the exception of PaCHS1, which exhibited high levels of constitutive expression, showed relatively lower expression levels but had variable spatial distributions. This finding revealed functional diversification of duplicate CHS genes and confirmed that gene duplication was followed by divergence. In addition to the previously mentioned anthocyanin accumulation, CHSs also play crucial roles in plant resistance, which indicates that CHS gene expression results from stimulation of abiotic or biotic stresses, such as UV light (Han et al., 2006), pathogens, low temperature and wounding (Dao et al., 2011). Stress-induced CHS expression levels have been reported in A. thaliana (Feinbaum and Ausubel, 1988; Leyva et al., 1995; Schenk et al., 2000; Wade et al., 2001), Hordeum vulgare (Christensen et al., 1998), A. majus (Junghans et al., 1993), Daucus carota (Glassgen et al., 1998), Brassica rapa (Zhou et al., 2007) and S. bicolor (Lue et al., 1989). Most of the Z. mays CHS genes were up- or down-regulated after salicylic acid treatment, which is associated with abiotic stresses (Han et al., 2016). In addition, the induction of CHS expression by the plant parasite, Orobanche aegyptiaca, which forms a physical connection with host roots, has been reported in diverse plant species (Griffitts et al., 2004). Of the five CHS genes, PaCHS1 is the most distant paralogue and is solely located on a single chromosome. The other four paralogous CHS genes were clustered in the Phalaenopsis-specific subclade of clade 1 (Fig. 4). In combination with chromosomal distributions, the four CHS genes were further classified into two groups, PaCHS2 and the gene cluster composed of PaCHS3, PaCHS4 and PaCHS5. The three genes were tandemly arrayed, which led us to speculate that expansion of the CHS gene family in P. aphrodite occurred through segmental duplications followed by tandem duplications (Fig. 3). In a previous study (Chomicki et al., 2015), the five homologous PhCHS genes in a Phalaenopsis hybrid cultivar included, instead of five paralogues, a mixture of paralogues and orthologues due to sexual hybridization of wild species (Fig. 4; Supplementary Data Fig. S3). We suggest studying gene functions in a gene family using a wild species to avoid redundancy of orthologues and missing paralogues. In addition, Cymbidium floriboundum CHS in that study seems to be clustered in our Phalaenopsis-specific subclade. The amino acid sequences of C. floriboundum CHSs are partially available (<58 % of full-length of PaCHSs; 223 vs. 387–94 of amino acids); therefore, we did not include C. floriboundum CHSs in our analysis.
In this study, we classified the five CHS genes into three groups that were located on three distinct chromosomes. Based on the expression patterns of the five CHS genes, the clustering was correlated with their sequence divergences and chromosomal rearrangements. This correlation has not been found in other plant gene families, such as the CHS gene family of Z. mays (Han et al., 2016) or the flavonol synthase gene family of A. thaliana (Owens et al., 2008).
SUPPLEMENTARY DATA
Supplementary data are available online at https://academic.oup.com/aob and consist of the following. Table S1: sequence information of CHSs used in the phylogenetic analysis. Table S2: sequences of primers used to amplify probe DNA for FISH mapping. Fig. S1: alignment of the amino acid sequences of the five CHSs identified in P. aphrodite. Fig. S2: chalcone synthase (CHS) phylogenetic relationships and their encoding gene structures in P. aphrodite. Fig. S3: phylogenetic relationships among the Phalaenopsis CHSs.
ACKNOWLEDGEMENTS
We thank Mallory Eckstut, PhD, from the Edanz Group (www.edanzediting.com/ac) for editing a draft of this manuscript. This work was supported by Innovative Translational Agriculture Research from Academia Sinica, Taiwan [2016AG001 and 2017PM001 to S.-B.C.].
LITERATURE CITED
- Arabidopsis Genome Initiative 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815. [DOI] [PubMed] [Google Scholar]
- Cai J, Liu X, Vanneste K, et al. . 2015. The genome sequence of the orchid Phalaenopsis equestris. Nature Genetics 47: 65–72. [DOI] [PubMed] [Google Scholar]
- Chao YT, Yen SH, Yeh JH, Chen WC, Shih MC. 2017. Orchidstra 2.0 A transcriptomics resource for the orchid family. Plant and Cell Physiology 58: e9. [DOI] [PubMed] [Google Scholar]
- Chomicki G, Bidel LPR, Ming F, et al. . 2015. The velamen protects photosynthetic orchid roots against UV-B damage, and a large dated phylogeny implies multiple gains and losses of this function during the Cenozoic. New Phytologist 205: 1330–1341. [DOI] [PubMed] [Google Scholar]
- Christensen AB, Gregersen PL, Schroder J, Collinge DB. 1998. A chalcone synthase with an unusual substrate preference is expressed in barley leaves in response to UV light and pathogen attack. Plant Molecular Biology 37: 849–857. [DOI] [PubMed] [Google Scholar]
- Dao TTH, Linthorst HJM, Verpoorte R. 2011. Chalcone synthase and its functions in plant resistance. Phytochemistry Reviews 10: 397–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X, Bashandy H, Ainasoja M, et al. . 2014. Functional diversification of duplicated chalcone synthase genes in anthocyanin biosynthesis of Gerbera hybrida. New Phytologist 201: 1469–1483. [DOI] [PubMed] [Google Scholar]
- Dressler RL. 1993. Phylogeny and classification of the orchid family. Cambridge: Cambridge University Press. [Google Scholar]
- Durbin ML, Learn GH, Huttley GA, Clegg MT. 1995. Evolution of the chalcone synthase gene family in the genus Ipomoea. Proceedings of the National Academy of Sciences, USA 92: 3338–3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durbin ML, McCaig B, Clegg MT. 2000. Molecular evolution of the chalcone synthase multigene family in the morning glory genome. Plant Molecular Biology 42: 79–92. [PubMed] [Google Scholar]
- Feinbaum RL, Ausubel FM. 1988. Transcriptional regulation of the Arabidopsis thaliana chalcone synthase gene. Molecular and Cellular Biology 8: 1985–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39: 783–791. [DOI] [PubMed] [Google Scholar]
- Ferrer JL, Jez JM, Bowman ME, Dixon RA, Noel JP. 1999. Structure of chalcone synthase and the molecular basis of plant polyketide biosynthesis. Nature Structural Biology 6: 775–784. [DOI] [PubMed] [Google Scholar]
- Ferreyra MLF, Rius SP, Casati P. 2012. Flavonoids: biosynthesis, biological functions, and biotechnological applications. Frontiers in Plant Science 3: 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franken P, Niesbachklosgen U, Weydemann U, Marechaldrouard L, Saedler H, Wienand U. 1991. The duplicated chalcone synthase genes C2 and Whp (white pollen) of Zea mays are independently regulated; evidence for translational control of Whp expression by the anthocyanin intensifying gene in. EMBO Journal 10: 2605–2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Givnish TJ, Spalink D, Ames M et al. . 2015. Orchid phylogenomics and multiple drivers of their extraordinary diversification. Proceedings of the Royal Society B: Biological Sciences 282: 171–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glassgen WE, Rose A, Madlung J, Koch W, Gleitz J, Seitz HU. 1998. Regulation of enzymes involved in anthocyanin biosynthesis in carrot cell cultures in response to treatment with ultraviolet light and fungal elicitors. Planta 204: 490–498. [DOI] [PubMed] [Google Scholar]
- Goto-Yamamoto N, Wan GH, Masaki K, Kobayashi S. 2002. Structure and transcription of three chalcone synthase genes of grapevine (Vitis vinifera). Plant Science 162: 867–872. [Google Scholar]
- Griffitts AA, Cramer CL, Westwood JH. 2004. Host gene expression in response to Egyptian broomrape (Orobanche aegyptiaca). Weed Science 52: 697–703. [Google Scholar]
- Han YH, Ding T, Su B, Jiang HY. 2016. Genome-wide identification, characterization and expression analysis of the chalcone synthase family in maize. International Journal of Molecular Sciences 17: 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han YY, Ming F, Wang W, Wang JW, Ye MM, Shen DL. 2006. Molecular evolution and functional specialization of chalcone synthase superfamily from Phalaenopsis Orchid. Genetica 128: 429–438. [DOI] [PubMed] [Google Scholar]
- Helariutta Y, Kotilainen M, Elomaa P, et al. . 1996, Duplication and functional divergence in the chalcone synthase gene family of Asteraceae: evolution with substrate change and catalytic simplification. Proceedings of the National Academy of Sciences, USA 93: 9033–9038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Hof K, van den Berg RG, Gravendeel B. 2008. Chalcone synthase gene lineage diversification confirms allopolyploid evolutionary relationships of European rostrate violets. Molecular Biology and Evolution 25: 2099–2108. [DOI] [PubMed] [Google Scholar]
- Hsu CC, Chen YY, Tsai WC, Chen WH, Chen HH. 2015. Three R2R3-MYB transcription factors regulate distinct floral pigmentation patterning in Phalaenopsis spp. Plant Physiology 168: 175–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu LF, He HH, Zhu CL, et al. . 2017. Genome-wide identification and phylogenetic analysis of the chalcone synthase gene family in rice. Journal of Plant Research 130: 95–105. [DOI] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project 2005. The map-based sequence of the rice genome. Nature 436: 793–800. [DOI] [PubMed] [Google Scholar]
- Ito M, Ichinose Y, Kato H, Shiraishi T, Yamada T. 1997. Molecular evolution and functional relevance of the chalcone synthase genes of pea. Molecular and General Genetics 255: 28–37. [DOI] [PubMed] [Google Scholar]
- Jander G, Barth C. 2007. Tandem gene arrays: a challenge for functional genomics. Trends in Plant Science 12: 203–210. [DOI] [PubMed] [Google Scholar]
- Junghans H, Dalkin K, Dixon RA. 1993. Stress responses in alfalfa (Medicago sativa L). 15. Characterization and expression patterns of members of a subset of the chalcone synthase multigene family. Plant Molecular Biology 22: 239–253. [DOI] [PubMed] [Google Scholar]
- Koduri PKH, Gordon GS, Barker EI, Colpitts CC, Ashton NW, Suh DY. 2010. Genome-wide analysis of the chalcone synthase superfamily genes of Physcomitrella patens. Plant Molecular Biology 72: 247–263. [DOI] [PubMed] [Google Scholar]
- Koes RE, Spelt CE, Vandenelzen PJM, Mol JNM. 1989. Cloning and molecular characterization of the chalcone synthase multigene family of Petunia hybrida. Gene 81: 245–257. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution 33: 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo YT, Hsu HL, Yeh CH, Chang SB. 2016. Application of a modified drop method for high-resolution pachytene chromosome spreads in two Phalaenopsis species. Molecular Cytogenetics 9: 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyva A, Jarillo JA, Salinas J, Martinezzapater JM. 1995. Low-temperature induces the accumulation of phenylalanine ammonia-lyase and chalcone synthase mRNAs of Arabidopsis thaliana in a light-dependent manner. Plant Physiology 108: 39–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li GQ, Moriyama EN. 2004. Vector NTI, a balanced all-in-one sequence analysis suite. Briefings in Bioinformatics 5: 378–388. [DOI] [PubMed] [Google Scholar]
- Liew CF, Goh CJ, Loh CS, Lim SH. 1998. Cloning and characterization of full-length cDNA clones encoding chalcone synthase from the orchid Bromheadia finlaysoniana. Plant Physiology and Biochemistry 36: 647–656. [Google Scholar]
- Lo C, Coolbaugh RC, Nicholson RL. 2002. Molecular characterization and in silico expression analysis of a chalcone synthase gene family in Sorghum bicolor. Physiological and Molecular Plant Pathology 61: 179–188. [Google Scholar]
- Lue WL, Kuhn D, Nicholson RL. 1989. Chalcone synthase activity in sorghum mesocotyls inoculated with Colletotrichum graminicola. Physiological and Molecular Plant Pathology 35: 413–422. [Google Scholar]
- Messing J, Bharti AK, Karlowski WM, et al. . 2004. Sequence composition and genome organization of maize. Proceedings of the National Academy of Sciences, USA 101: 14349–14354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metsalu T, Vilo J. 2015. ClustVis: a web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Research 43: W566–W570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ober D. 2005. Seeing double: gene duplication and diversification in plant secondary metabolism. Trends in Plant Science 10: 444–449. [DOI] [PubMed] [Google Scholar]
- Owens DK, Alerding AB, Crosby KC, Bandara AB, Westwood JH, Winkel BSJ. 2008. Functional analysis of a predicted flavonol synthase gene family in Arabidopsis. Plant Physiology 147: 1046–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchy N, Lehti-Shiu M, Shiu SH. 2016. Evolution of gene duplication in plants. Plant Physiology 171: 2294–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitakdantham W, Sutabutra T, Chiemsombat P, Pitaksutheepong C. 2010. Isolation and characterization of chalcone synthase gene isolated from Dendrobium Sonia Earsakul. Pakistan Journal of Biological Sciences 13: 1000–1005. [DOI] [PubMed] [Google Scholar]
- Rizzon C, Ponger L, Gaut BS. 2006. Striking similarities in the genomic distribution of tandemly arrayed genes in Arabidopsis and rice. PLoS Computational Biology 2: 989–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M. 1987. The neighbor–joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution 4: 406–425. [DOI] [PubMed] [Google Scholar]
- Saslowsky DE, Dana CD, Winkel-Shirley B. 2000. An allelic series for the chalcone synthase locus in Arabidopsis. Gene 255: 127–138. [DOI] [PubMed] [Google Scholar]
- Schenk PM, Kazan K, Wilson I, et al. . 2000. Coordinated plant defense responses in Arabidopsis revealed by microarray analysis. Proceedings of the National Academy of Sciences, USA 97: 11655–11660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SD, Rausher MD. 2011. Gene loss and parallel evolution contribute to species difference in flower color. Molecular Biology and Evolution 28: 2799–2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SD, Wang S, Rausher MD. 2013. Functional evolution of an anthocyanin pathway enzyme during a flower color transition. Molecular Biology Evolution 30: 602–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer H, Saedler H. 1986. Structure of the chalcone synthase gene of Antirrhinum majus. Molecular and General Genetics 202: 429–434. [DOI] [PubMed] [Google Scholar]
- Su CL, Chao YT, Yen SH, et al. . 2013. Orchidstra: an integrated orchid functional genomics database. Plant and Cell Physiology 54: e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. 1994. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai WC, Fu CH, Hsiao YY, et al. . 2013. OrchidBase 2.0: comprehensive collection of Orchidaceae floral transcriptomes. Plant and Cell Physiology 54: e7. [DOI] [PubMed] [Google Scholar]
- Tuskan GA, Difazio S, Jansson S, et al. . 2006. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313: 1596–1604. [DOI] [PubMed] [Google Scholar]
- Wade HK, Bibikova TN, Valentine WJ, Jenkins GI. 2001. Interactions within a network of phytochrome, cryptochrome and UV-B phototransduction pathways regulate chalcone synthase gene expression in Arabidopsis leaf tissue. The Plant Journal 25: 675–685. [DOI] [PubMed] [Google Scholar]
- Yang J, Huang JX, Gu HY, Zhong Y, Yang ZH. 2002. Duplication and adaptive evolution of the chalcone synthase genes of Dendranthema (Asteraceae). Molecular Biology and Evolution 19: 1752–1759. [DOI] [PubMed] [Google Scholar]
- Zavala K, Opazo JC. 2015. Lineage-specific expansion of the chalcone synthase gene family in rosids. PLoS One 10: e0133400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong XB, de Jong JH, Zabel P. 1996. Preparation of tomato meiotic pachytene and mitotic metaphase chromosomes suitable for fluorescence in situ hybridization (FISH). Chromosome Research 4: 24–28. [DOI] [PubMed] [Google Scholar]
- Zhou B, Li YH, Xu ZR, Yan HF, Homma S, Kawabata S. 2007. Ultraviolet A-specific induction of anthocyanin blosynthesis in the swollen hypocotyls of turnip (Brassica rapa). Journal of Experimental Botany 58: 1771–1781. [DOI] [PubMed] [Google Scholar]
- Zuckerkandl E, Pauling L. 1965. Evolutionary divergence and convergence in proteins. Evolving genes and proteins. New York: Academic Press, 97–166. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.