Figure 4.
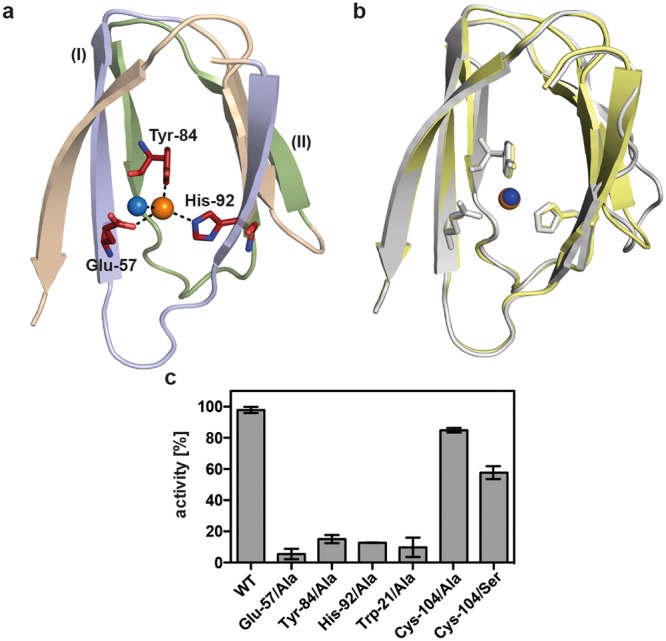
The iron-binding site in the (Pl)EctC protein and enzyme activities of selected (Pl)EctC variants. (a) The iron (represented by an orange sphere) is coordinated by the side-chains of Glu-57, Tyr-84, and His-92 of the (Pl)EctC protein with distances of 2.9 Å, 2.8 Å, and 2.9 Å, respectively. The iron-binding site in the substrate-free (Pl)EctC crystal structure also contains a localized water molecule (blue sphere); it has a distance of 2.9 Å to the iron atom. The two conserved cupin-motifs include those residues that coordinate the metal ion [G(X)5WY(X)4E(X)6G; G(X)6PG(X)2Y(X)3G(X)3H; letters in bold indicate metal-binding residues] are highlighted as part of the overall protein (Pl)EctC crystal structure. The first cupin motif [G(X)5WY(X)4E(X)6G] is shown in blue, and the second cupin motif [G(X)6PG(X)2Y(X)3G(X)3H] is represented in green. A number of secondary structure elements of the (Pl)EctC protein were removed in order to highlight the architecture of the iron-binding site and the position of the two cupin motifs. (b) Overlay of the iron-binding site in the (Pl)EctC (shown in yellow) and (Sa)EctC (shown in grey) crystal structures. The three residues involved in the binding of the Fe(II) ion are depicted as sticks. A water molecule (blue sphere) in the (Sa)EctC crystal structure occupies the same location as the Fe(II) ion (orange sphere) in the (Pl)EctC structure. (c) Single amino acid substitution variants of the (Pl)EctC protein were assayed for their enzyme activity. Enzyme activity measurements were conducted under buffer conditions [20 mM HEPES buffer (pH 8.5), 50 mM NaCl, 0.1 mM (NH4)2Fe(SO4)2] optimized for the wild-type enzyme using 10 µg of protein and 10 mM of the substrate N-γ-ADABA. The enzyme assays were conducted at 30 °C and run for 30 min and the formation of ectoine was monitored by HPLC analysis. The enzyme activity of the mutant (Pl)EctC proteins is represented relative to that of the wild-type enzyme (set at 100% activity).
