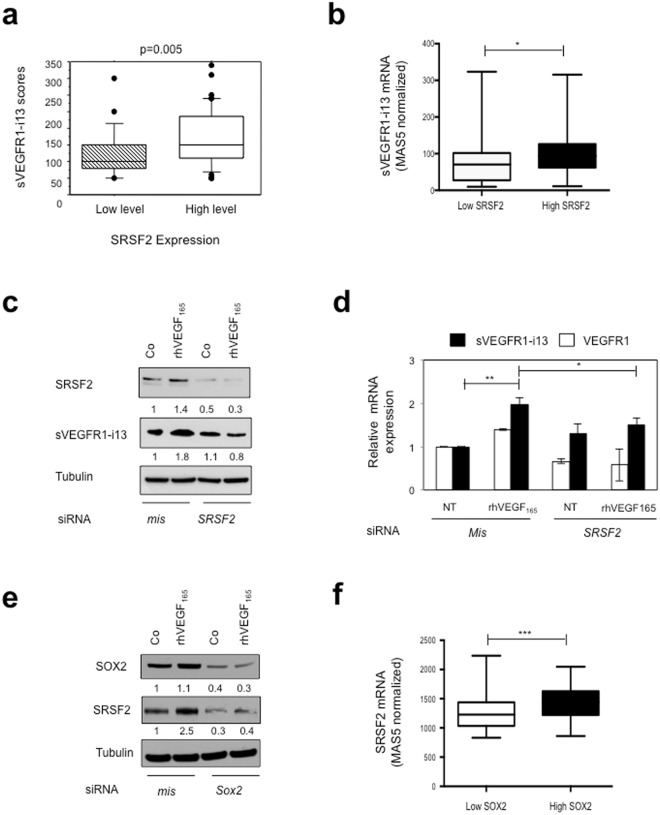
Figure 3.
SRSF2 cooperates with SOX2 to regulate sVEGFR1-i13 expression in SQLC cells. (a) Mean levels ± SD of sVEGFR1-i13 immunohistochemical scores according to the SRSF2 status in squamous cell lung carcinoma, where SQLC are sub-divided in two classes representing tumors with high or low levels of SRSF2 compared to normal lung tissues4,24. (b) Mean levels ± SD of MAS5-normalized sVEGFR1-i13 mRNA in SQLC patients taken from the GSE4573 database expressing either low (<50th percentile) or high (>50th percentile) levels of SRSF2 mRNA. (c,d) MGH7 cells were transfected with mismatch (mis) or Srsf2 (SRSF2) siRNA during 48 hours and treated or not (Co) for 24 additional hours with 1 ng/ml rhVEGF165. (c) Western blot analyses of SRSF2 and sVEGFR1-i13 proteins. Tubulin was used as a loading control. Numbers represent the quantification of SOX2, sVEGFR1-i13 or VEGF165 signal intensities using Image J software. The value 1 was arbitrarily assigned to the untreated condition signal. (d) RT-qPCR analyses of sVEGFR1-i13 and VEGFR1. GAPDH was used as an internal control. (e) Western blot analyses of SOX2 and SRSF2 proteins in MGH7 cells transfected with mismatch (mis) or Sox2 (Sox2) siRNA during 48 hours and treated or not (Co) for 24 additional hours with 1 ng/ml rhVEGF165. Tubulin was used as a loading control. Quantification as in (c). (f) Mean levels ± SD of MAS5-normalized Srsf2 mRNA in SQLC patients taken from the GSE4573 database expressing either low (<50th percentile) or high (>50th percentile) levels of Sox2 mRNA. Statistical analyses were performed using a non parametric Mann-Whitney test (*p < 0.05; **p < 0.01; ***p < 0.001). All western blot experiments were performed at least three times. Illustrations of a representative result are presented for each condition.