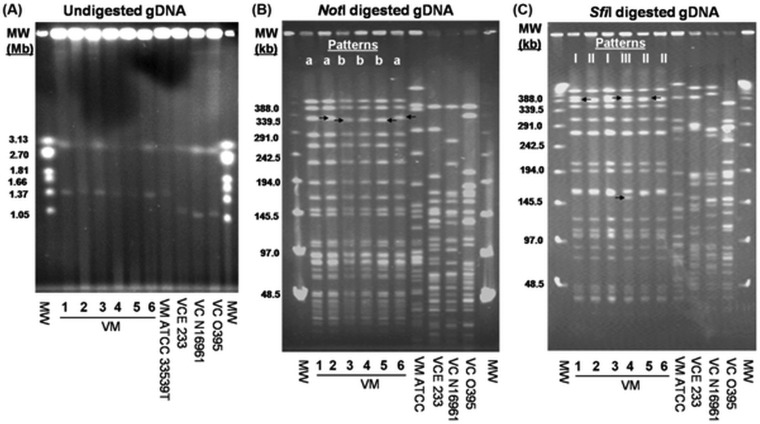
FIG 1.
PFGE analysis of environmental ctx+ and reference (ATCC) strains of V. mimicus (VM) and of ctx+ non-O1/non-O139 (VCE 233), O1 El Tor (VC N16961), and O1 classical (VC O395) strains of V. cholerae. (A) Gel image of PFGE profiles of undigested gDNA showing similar sizes of the two chromosomes of ctx+ V. mimicus and ATCC V. mimicus strains. (B and C) Gel images showing PFGE patterns of NotI-digested (B) and SfiI-digested (C) gDNA of ctx+ V. mimicus and the reference strains. The ctx+ V. mimicus strains were clonal but differed in 1 to 2 bands, indicated by arrows. NotI- and SfiI-digested gDNA of ctx+ V. mimicus strains generated two (a and b) and three (I, II, and III) PFGE profiles, respectively. “MW” represents the molecular weight standard of the Hansenula wingei chromosomes (Bio-Rad) for undigested gDNA (A) and lambda ladder (Bio-Rad) for digested gDNA (B and C).