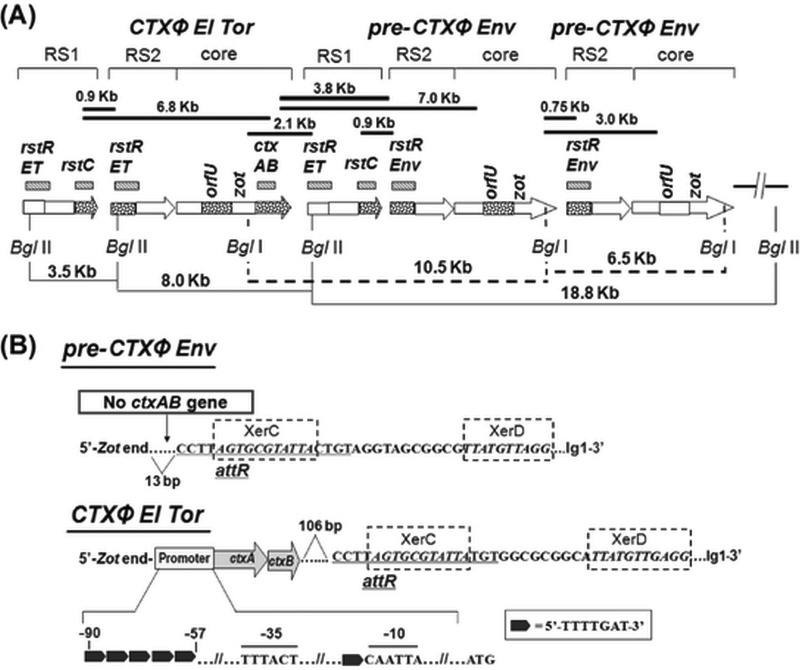
FIG 3.
Organization of CTXΦEl Tor, RS1, and pre-CTXΦEnv in V. mimicus strains. (A) Filled bars indicate the PCR arrays used to check probable locations of genes, and the sizes of PCR products are given at the top. Hashed bars indicate the genetic regions (names mentioned at the top) used as probes for Southern hybridization after restriction digestion with BglI or BglII enzymes; arrows indicate RS1, RS2, or core prophage data where dotted regions were analyzed by sequencing. Lines (filled and dotted) at the bottom show the distances between specific genetic locations determined by Southern hybridization analysis of the BglI- or BglII-digested genomic DNA performed using specific probes. (B) Signature sequences in the flanking region of pre-CTXΦEnv and CTXΦEl Tor in V. mimicus. The ctxAB promoter of CTXΦEl Tor contains 5 heptamer (TTTTGAT) repeats, shown by filled black arrows, which is characteristic of classical type ctxAB. The phage integration site of the CTXΦEl Tor in V. mimicus, the attR sequence, followed by XerC and XerD binding sites, starts at 106 bp downstream of ctxAB gene, which is similar to the reference El Tor strain, N16961, of V. cholerae. However, the phage integration site, characterized by these signature sequences, of pre-CTXΦEnv in V. mimicus starts at 13 bp downstream of the zot gene.