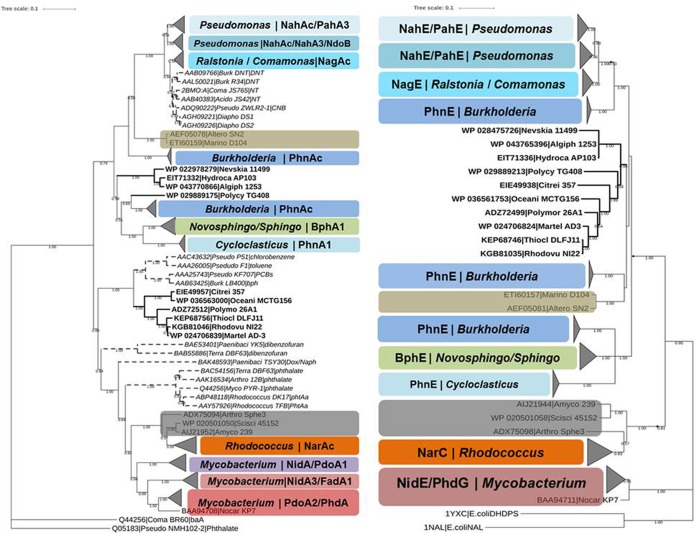
FIG 3.
Comparison of PahE and PahAc-based phylogenies of PAH-degrading bacteria. Both panels show majority rule consensus trees based on the neighbor-joining (NJ) method. Bootstrap support is indicated at individual branches, and only values greater than 0.5 are shown. Different types of both genes are marked by different colors. The same color in both trees indicates a one-to-one correspondence in both genes and that the two genes are from same degrading bacteria. The bold text in both trees indicates members of the group of newly defined potential PAH degraders, which also correspond to each other. The other aromatic ring-hydroxylating dioxygenase clades (other-ArhAc, which could not degrade PAH) in PahAc trees are indicated by dashed branches. The size of node triangles indicates the number of sequences in each clade. The scale bar indicates 0.1 branch distance. Accession numbers are shown to the left of each organism name.