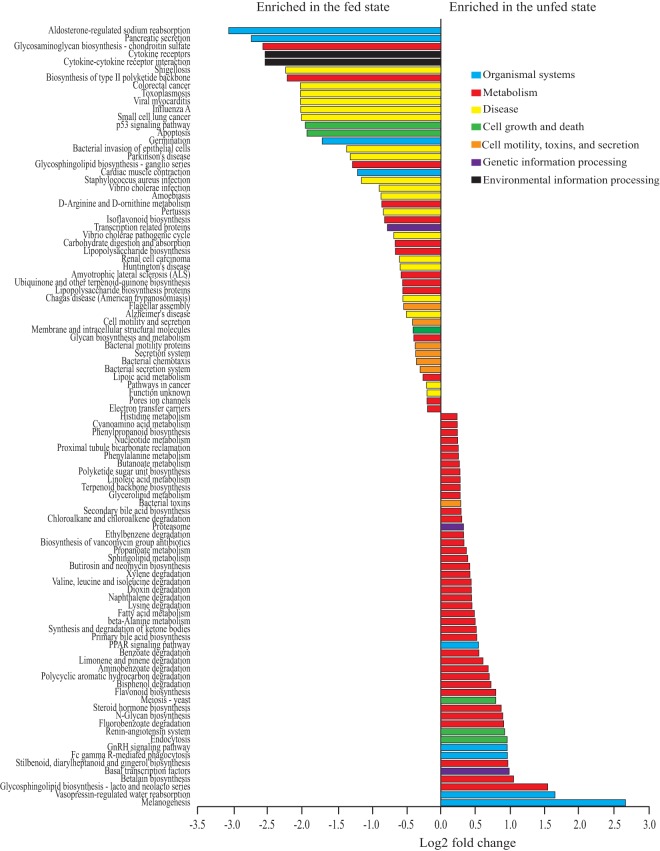
FIG 4.
Predicted functional gene categories vary significantly between unfed-state (1100 and 2000 h) and fed-state (1230, 1400, and 1600 h) time points, with fed-state time points enriched in disease-associated pathways and unfed-state time points enriched in diverse metabolic pathways. Metagenomes were predicted from amplicon data using PICRUSt and the third hierarchical level of KEGG. Differential abundance was evaluated using DESeq2 in R, with gene categories having an adjusted P value of <0.05 shown here. Colors represent larger subsystem categories in KEGG. Only the top 99 of 111 significant pathways are plotted (n = 17 fish at 1100 and 2000 h and 18 fish at 1230, 1400, and 1600 h).