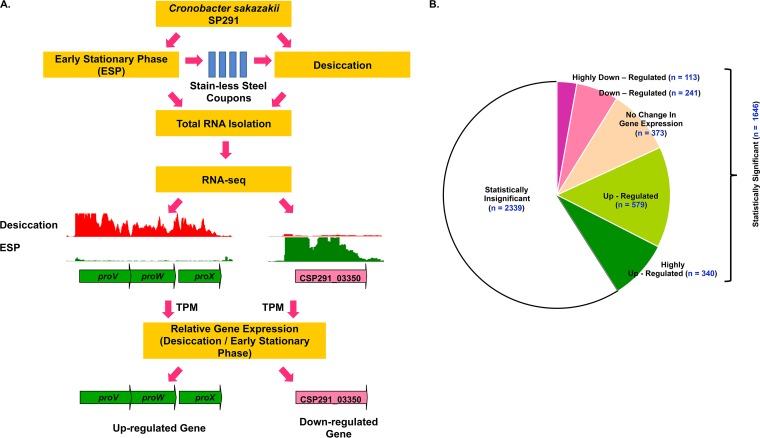
FIG 3.
(A) A figure summarizing the experimental strategy deployed for the RNA-seq experiment. ESP-grown C. sakazakii SP291 cells were subjected to desiccation for 4 h on sterile stainless steel coupons, and total RNA was purified from both ESP-grown and desiccated cells. The graphs represent the RNA-seq reads mapped uniquely against the C. sakazakii SP291 genome in the respective condition, as visualized in Integrated Genome Browser. The genes on the C. sakazakii SP291 genome were categorized as upregulated (green arrows) or downregulated (pink arrows) based on the ratio of gene expression values, desiccation versus ESP. (B) Number of differentially regulated genes in desiccated C. sakazakii SP291 (Table S1, WS2). The differential expression was calculated from the ratio of TPM-based gene expression data, desiccation versus ESP. The thresholds for the selection of upregulated and downregulated genes are mentioned in Materials and Methods. TPM, transcripts per million approach.