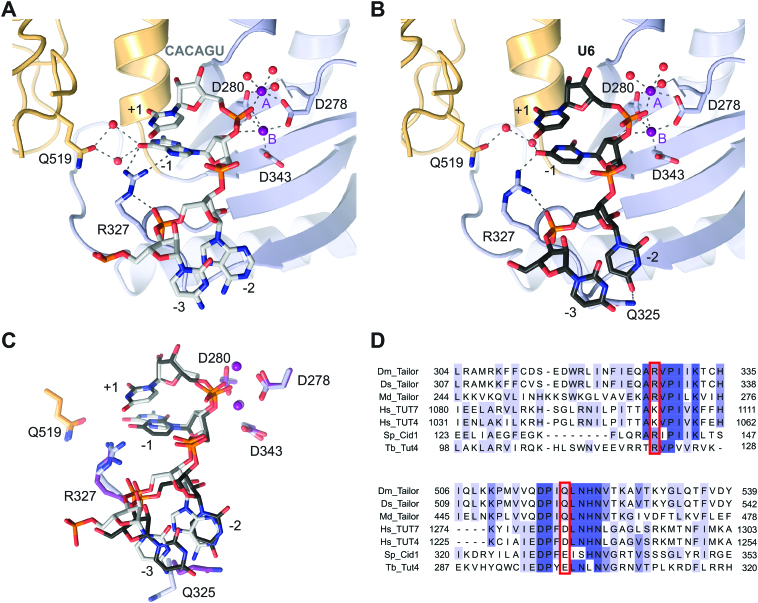
Figure 2.
Structures of product-bound DmTailor complexes. (A) Zoom-in view of the active site of DmTailor bound to a CACAGU hexanucleotide (grey sticks) and two Mg2+ ions (purple spheres). DmTailor lobes are colored as in Figure 1. The catalytic aspartates and the Arg327 and Gln519 residues are shown in stick format, hydrogen bonds and metal coordination bonds are shown as grey dashed lines, water molecules are shown as red spheres. (B) Zoom-in view of the active site of DmTailor bound to a U6 hexanucleotide (black sticks) and two Mg2+ ions (purple spheres). (C) Superposition of the CACAGU and U6 complex structures in (A) and (B). The structures were superimposed using DALI (48). The structures are colored as in (A) and (B) with the residues and magnesium atoms of the U6-bound DmTailor in darker colors. (D) Sequence alignment of Drosophila melanogaster Tailor (Dm_Tailor), Drosophila sechellia Tailor (Ds_Tailor), Musca domestica Tailor (Md_Tailor), Homo sapiens TUT7 (Hs_TUT7), Homo sapiens TUT4 (Hs_TUT4), Schizosaccharomyces pombe Cid1 (Sp_Cid1) and Trypanosoma brucei Tut4 (Tb_Tut4). The sequences were aligned using MAFFT version 7 (51). Conserved residues are highlighted in darker shades of blue with increasing degree of conservation (invariant in dark blue). Putative residues involved in acceptor substrate specificity, Arg327 and Gln519, are highlighted using red boxes. For a sequence alignment of the full nucleotidyl transferase domain, see Supplementary Figure S4.