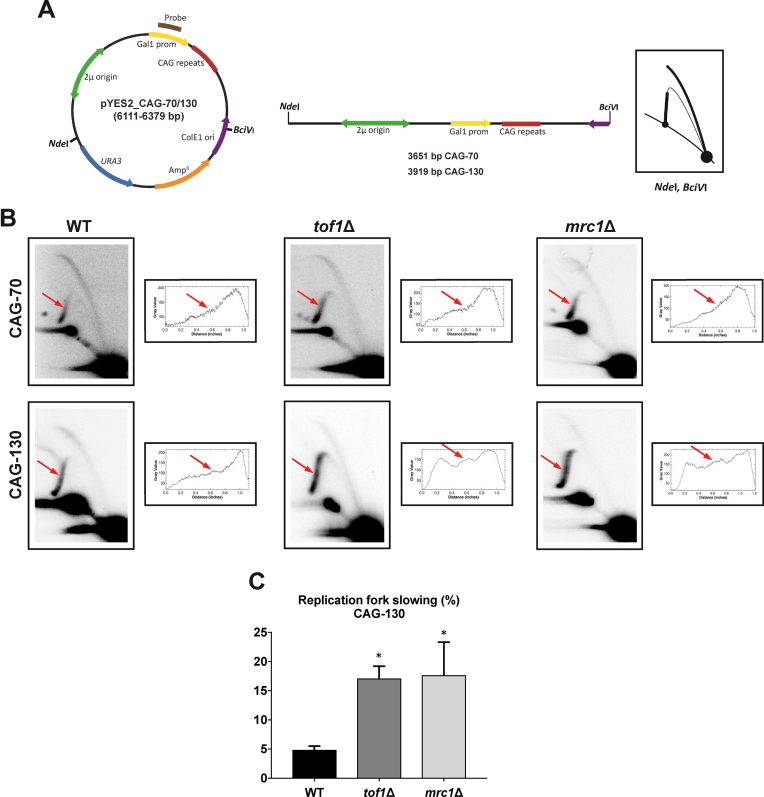
Figure 2.
Analysis of replication through CAG-70 and CAG-130 repeats by two-dimensional (2D) agarose gel electrophoresis in WT, tof1Δ and mrc1Δ strains. (A) Schematic of the pYES2 constructs is shown with its mass and genetic map. The relative positions of its most relevant features are indicated inside: the 2 μm origin, the ColE1 unidirectional origin (ColE1 Ori), the ampicillin-resistance gene (AmpR), URA3, the GAL1 promoter (Gal1 prom) and 70 or 130 CAG repeats. Outside, the relative positions of sites recognized by the restriction endonucleases NdeI and BciVI are indicated. To the right, is shown the corresponding linear map of the pYES2 plasmid restriction fragment with the sizes and the diagrammatic interpretation if replication initiates bi-directionally at the 2 μm origin and proceeds unconstrained. (B) Representative 2D gels of replication through CAG-70 and CAG-130 repeats in WT, tof1Δ, and mrc1Δ strains. DNA was isolated, digested with NdeI and BciVI and analyzed by 2D gel. Red arrow points to the location of the CAG repeats. To the right of each 2D gel are shown the densitometric profiles corresponding to the Y-arc region where the (CAG)n repeats are located; peaks on densitograms correspond to bulges on the Y-arcs. A representative gel and its corresponding profile is shown; three experiments were analyzed for each strain. (C) Quantification of replication fork slowing in pYES2 CAG-130 in WT, tof1Δ and mrc1Δ strains. The ratio of radioactivity in the peak area to that corresponding area of a smooth replication arc reflects the extent of replication slowing. Three different experiments were performed for each strain. Percentage of replication fork slowing is 3.3%, 4.6% and 5.8% for WT, 13.2%, 17.7% and 20.4% for tof1Δ, and 8.3%, 16.4% and 27.7% for mrc1Δ. Error bars indicate standard error of the mean. The star indicates a significant difference between wild-type and mutants. P = 0.0483 (tof1Δ versus WT), P = 0.0378 (mrc1Δ versus WT).