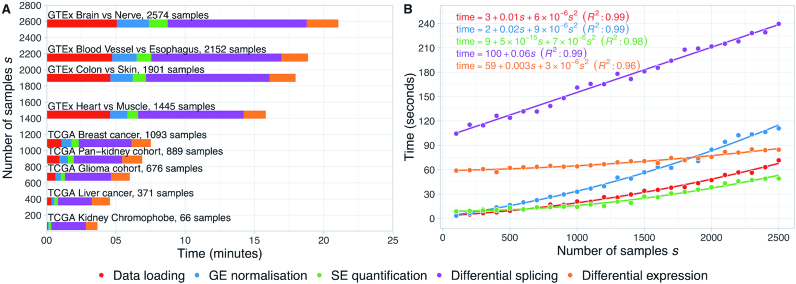
Figure 3.
Performance benchmark for alternative splicing analysis using RNA-seq data from multiple TCGA and GTEx sample types. (A) Median times of 10 runs of data loading, gene expression (GE) normalisation, skipped exon (SE) event quantification and differential expression and splicing analysis (normal versus tumour for TCGA data or pairwise tissue comparison for GTEx data) using psichomics. The default settings were used during the runs. (B) Estimation of the time complexity of each of the aforementioned steps in psichomics. Randomly generated synthetic datasets of different sample size s were used as input. Equations and coefficient of determination (R2) for the best fits are displayed.