Figure 1.
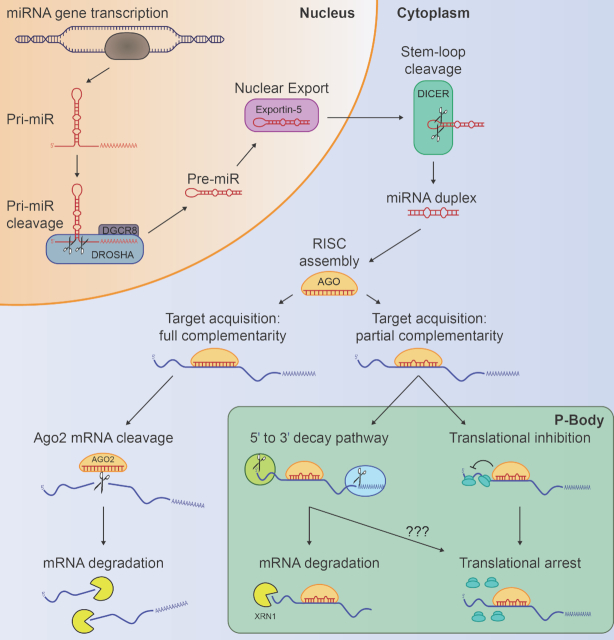
MicroRNA biogenesis and mechanisms of transcript repression. miRNA host genes are transcribed by RNA polymerase II, yielding primary miRNA (pri-miR) transcripts containing hairpin secondary structures (20). These ∼70nt hairpins, known as precursor miRNAs (pre-miRs), are liberated from pri-miRs by the Drosha-DGCR8 nuclear microprocessor complex (21) and then exported to the cytoplasm through nuclear pores upon association with Exportin-5 (22,23). In the cytoplasm, RNase III Dicer cleaves the hairpin loop to produce a double stranded miRNA duplex (24), from which one strand is loaded into an Argonaute (Ago) family protein (25–27). Full base complementarity between a miRNA associated with Ago2 and target mRNA results in direct endonucleolytic cleavage of the transcript by Ago2, followed by degradation (33). Partial miRNA-mRNA complementarity triggers either 5′ to 3′ mRNA decay—with potential for translational arrest—(30,31,37–41), or direct inhibition of mRNA translation (44–49,52,53,55,56), both of which are thought to be localized with subcellular P-bodies (62,63).