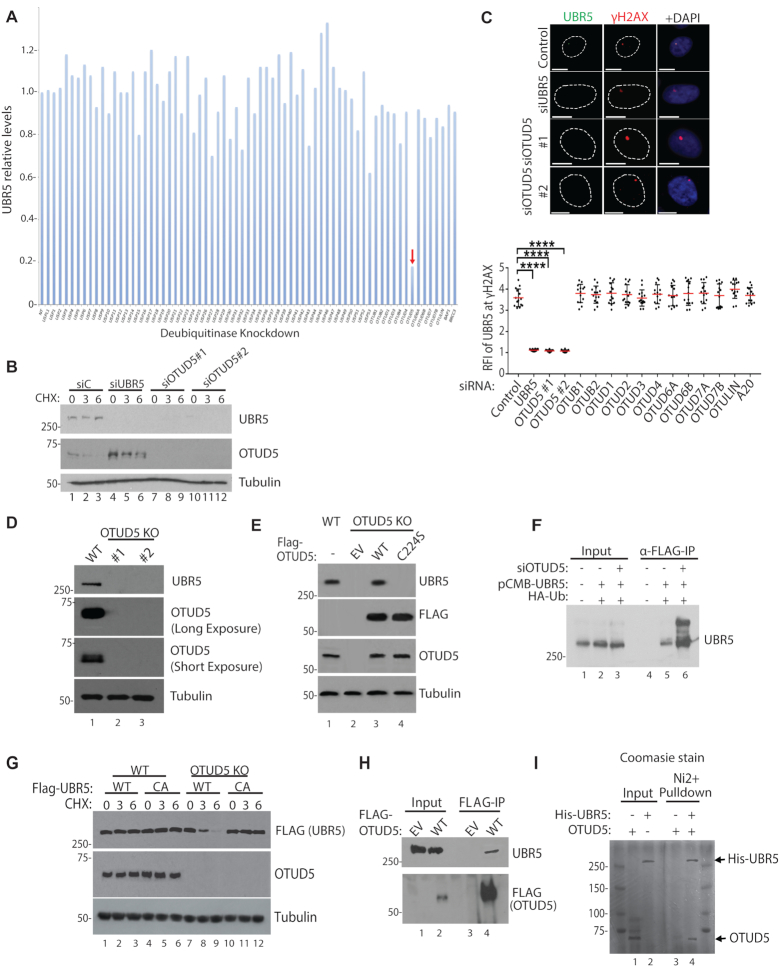
Figure 1.
OTUD5 is a specific stabilizer of UBR5. (A) Indicated siRNAs (20 nM) were transfected to 293T cells, pellets were harvested after 72 hours and UBR5 levels were detected by western blots. Band intensities were internally normalized to tubulin and quantified using Image J. (B) siRNAs were transfected to HeLa, followed by Cycloheximide treatment (10 µM, for indicated hours) and western blotting. (C) UBR5 foci formation was induced by UVC through 3 µm micropore filter (100J/m2, 1 hour recovery) following the siRNA transfections (n = 20 each). See ‘Materials and Method’ section for RFI description. Bottom panel is for testing various (OTU DUB members) siRNAs for UBR5 foci formation (n = 20 each, **** indicates P-value < 0.0005). (D) Confirmation of OTUD5 CRISPR KO HeLa clones (#1 is CRISPR-Cas9 clone, #2 is Double-Nickase clone). (E) OTUD5 KO#1 cells were transfected with WT or C224S OTUD5 plasmids, and analyzed by western blotting. (F) 293T cells were transfected with indicated siRNAs and the plasmids, harvested pellets were lysed and anti-FLAG IP assay was performed. Bands above unmodified UBR5 are increased in OTUD5 Knockdown cells. (G) Cyclohexamide chase analysis (10 µM, for indicated hours) in OTUD5 KO cells complemented with either WT or catalytically inactive C2768A FLAG-UBR5 plasmids. (H) 293T cells were transfected with 3xFLAG-OTUD5 plasmid and anti-FLAG IP was performed. (I) Purified recombinant OTUD5 and 6xHis-UBR5 proteins were mixed, and the mixture was subject to Ni beads pulldown. In Ni2+ -only, OTUD5 proteins were added without UBR5. Shown is the coomassie stained gel.