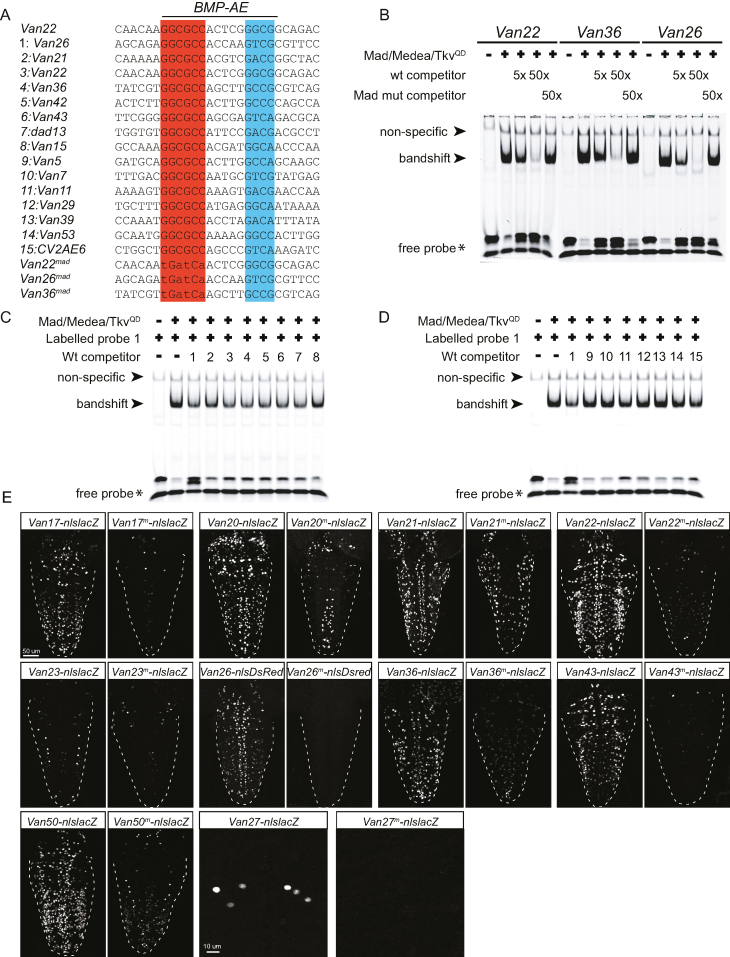
Figure 3.
The BMP-AE of wit-responsive genomic fragments is bound by pMad/Medea complexes with high affinity in vitro and is required in vivo. (A–D) Electrophoretic mobility shift assays (EMSA) for three selected BMP-AEs with S2 cell lysates transfected with Mad, Medea and TkvQD to activate BMP signaling. (A) DNA sequences used for EMSA analysis, highlighting the presumptive pMad (red) and Medea (blue) binding sites of the BMP-AE. The DNA sequences shown were taken from the predicted BMP-AE within the indicated genomic fragments. The BMP-AEs within Van22, Van36 and Van26 were used as IRDye700-labeled probes, as well as for unlabeled wildtype and mutated (Van36mad, Van26mad, Van22mad) competitors. All other sequences were used as unlabeled wildtype competitors. Mutant competitors were all mutated at the putative pMad-binding site. (B) EMSA with lysates from S2 cells transfected with the indicated plasmids (Mad/Medea/TkvQD) were incubated with the indicated IRDye700-labeled wildtype probes and run on non-denaturing gels. A band shift of each labeled probe was generated only in lanes loaded with S2 cell lysate transfected with Mad/Medea/TkvQD. Labeled probes were mixed with 5x or 50x stoichiometric excess of wildtype competitor (wt) or mutated (Mad mut) competitor. Five and 50x excess of wildtype competitor progressively diminished the band shift of the labeled probe, indicative of pMad/Medea complexes binding to the unlabeled competitor. In contrast, addition of the mutated competitor at a 50x excess resulted in retention of the labeled band shift, indicating that mutated competitors did not bind pMad/Medea complexes. (C, D) We tested the ability of fourteen BMP-AEs from wit-responsive (1–8 in A) and wit-non responsive (9–15 in A) genomic fragments to compete for pMad/Medea binding, as unlabeled DNA probes, with an IRDye700-labeled DNA probe for the BMP-AE of Van26. EMSA with lysates from S2 cells transfected with the indicated plasmids (Mad/Medea/TkvQD) were incubated with the IRDye700-labeled Van26 probe and the fourteen BMP-AE DNA unlabeled competitors. The labeled probe was mixed with a 5× stoichiometric excess of unlabeled competitors. The 5× excess of Van26 competitor diminished the band shift of the labeled probe, indicative of pMad/Medea complexes binding to the unlabeled competitor. The 5× excess of functional BMP-AEs within Van21, Van22, Van36, Van42, Van43, dad13 and Van15 competitors also resulted in a diminution of the band shift of the labeled probe (C). In contrast, the 5x excess of non-functional BMP-AEs within Van5, Van7, Van29, Van39 and Van53 did not result in a diminution of the band shift of the labeled probe (D), indicating that their affinity for the pMad/Medea complexes is lower than for the functional BMP-AEs within wit-responsive fragments. (E) Nuclear β-galactosidase and nlsDsred expression driven from 10 select wit-responsive fragments in which the BMP-AE motif was either wildtype or mutated at the Mad site. The point mutations that were introduced into the Mad site of the BMP-AEs are similar to the ones that abolished competition in vitro. In all cases, there was a reduction in the number of nuclei that expressed the nlsLacZ and nlsDsred reporters.