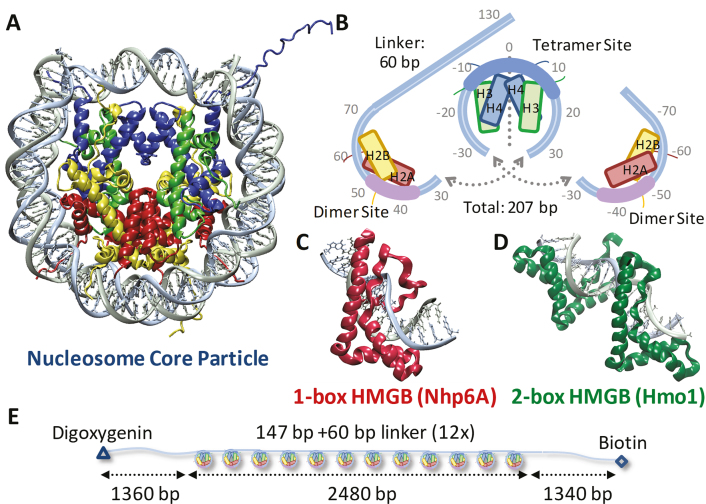
Figure 1.
Experiments probe histone–DNA interactions. (A) DNA wraps ∼1.7 left-handed turns around the histone octamer. (PDB code: 1aoi) (B) Nucleosome arrays consist of twelve repeats of this 11-nm diameter core particle (147 bp) and adjacent 20 nm long (60 bp) linkers. The bases are numbered from the dyad axis (dotted line) in this split image, which stacks vertically from left (top) to right (bottom). Helix-strand-helix structures of individual histones are represented by boxes aligned with each strand. At the dyad axis, the (H3–H4)2 tetramer binds up to 80 base pairs as the central region of DNA bound most strongly is shown in cyan. Off-dyad bases of the outer turns are bound to the H2A–H2B dimers across interactions distributed over the whole length of the outer turn DNA where the bases bound most strongly are highlighted in pink (26,47). (C) S. cerevisiae protein Nhp6A consists of a single L-shaped HMGB domain with an unstructured cationic N-terminus. (PDB code: 1j5n) (8) (D) NMR structure of recombinant HMGB protein thought to resemble S. cerevisiae Hmo1 bound to DNA, drawn to scale with (A) and (C). (PDB code: 2gzk) (7) Two HMGB domains (the second is in the foreground) are connected by a linker and flanked by a C-terminal domain believed to facilitate protein dimerization (not shown). Both Hmo1 and Nhp6A engage the DNA minor groove inserting the aromatic residues between its base pairs, thereby inducing strong bending in the helical axis of ∼90° toward the major groove (9,10). Unstructured tails are thought to play a role in charge stabilization inside that bend. (E) Arrays for AFM and OT experiments consist of twelve repeats of the 147-bp Widom 601 sequence with 60-bp linker, flanked by two ∼1350-bp non-nucleosomal sequences derived from plasmid pUC19 to serve as handles, labeled for attachment.