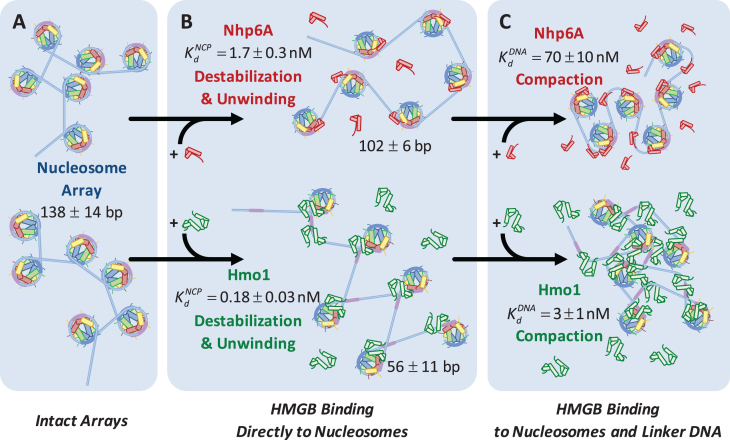
Figure 6.
HMGB induces nucleosome disruption and compaction. (A) For a small 6x ‘array’ in the absence of HMGB proteins (from Figure 1), sequential core particles are shown separated by linker DNA. The variability of linker length due to unwrapping and the possibility of nucleosome flipping leads to an irregularly dispersed 2D array shown. This follows the predictions of the model of Supplementary Methods 2. (B, upper path) Single box Nhp6A binds to the core particle (c ∼ 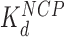 ), disrupting most of the DNA-(H2A–H2B) binding while DNA-(H3–H4)2 interactions remain intact. This leads to nucleosome dispersion, as the core particles appear further apart due to the released lengths of DNA (drawn roughly to scale using the numbers of Table 1). Though HMGB-nucleosome binding may be nearly saturated, there is little binding to double stranded DNA. (C, upper path) At protein concentrations approaching
), disrupting most of the DNA-(H2A–H2B) binding while DNA-(H3–H4)2 interactions remain intact. This leads to nucleosome dispersion, as the core particles appear further apart due to the released lengths of DNA (drawn roughly to scale using the numbers of Table 1). Though HMGB-nucleosome binding may be nearly saturated, there is little binding to double stranded DNA. (C, upper path) At protein concentrations approaching 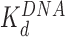 , protein binding induces DNA bending and random nucleosome compaction. (B, lower path) Double box Hmo1 binds to core particles (c ∼
, protein binding induces DNA bending and random nucleosome compaction. (B, lower path) Double box Hmo1 binds to core particles (c ∼ 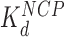 ), causing complete disruption of DNA-(H2A–H2B) binding and now some disruption of DNA-(H3–H4)2 interactions. This leads to the release of nearly half of the inner turn DNA, further dispersing the core particles of the array. Though it is not shown here, complete dissociation of the octamer is seen for 2–3 of the twelve nucleosomes at the peak of array dispersion. (C, lower path) Higher protein concentrations yield increased binding to linker DNA (c ∼
), causing complete disruption of DNA-(H2A–H2B) binding and now some disruption of DNA-(H3–H4)2 interactions. This leads to the release of nearly half of the inner turn DNA, further dispersing the core particles of the array. Though it is not shown here, complete dissociation of the octamer is seen for 2–3 of the twelve nucleosomes at the peak of array dispersion. (C, lower path) Higher protein concentrations yield increased binding to linker DNA (c ∼ 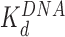 >>
>> 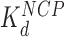 ), inducing nucleosome compaction. Numbers are averages of AFM and OT experiments, where appropriate. See Table 1 for full results and uncertainties.
), inducing nucleosome compaction. Numbers are averages of AFM and OT experiments, where appropriate. See Table 1 for full results and uncertainties.