Table 1.
Results from AFM and OT experiments on nucleosome arrays
| Technique | DNA per Nucleosomea (bp) | Nucleosomes per Arrayb (N/n) |
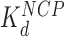 c (nM)
c (nM) |
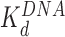 d (nM)
d (nM) |
|
|---|---|---|---|---|---|
| Arrays | AFM | 139 ± 14 | 11.6 ± 0.3 | – | – |
| OT | 74 ± 7 | 12.0 ± 0.2 | – | – | |
| Average | 139 ± 14 | 11.8 ± 0.2 | – | – | |
| Arrays +Nhp6A | AFM | 102 ± 6 | 10.2 ± 0.4 | 1.8 ± 0.6 | – |
| OT | 69 ± 9 | 11.0 ± 0.5 | 1.6 ± 0.2 | – | |
| Average | 102 ± 6 | 10.6 ± 0.3 | 1.7 ± 0.3 | 70 ± 10 | |
| Arrays +Hmo1 | AFM | 58 ± 20 | 9.8 ± 1.0 | 0.09 ± 0.04 | – |
| OT | 55 ± 7 | 8.0 ± 0.5 | 0.29 ± 0.05 | – | |
| Average | 56 ± 11 | 8.9 ± 0.6 | 0.18 ± 0.03 | 3 ± 1 |
AFM center-to-center length measurements and inner turn (tetrameric) lengths from OT experiments agree very well. For Hmo1 and Nhp6A, the AFM data were taken for concentrations where dispersion was maximized, while the protein concentrations in OT experiments were selected to assure binding saturation. See Methods for more information.
aDNA per nucleosome may be calculated in AFM and OT experiments. In AFM experiments, this is deduced from the linker DNA lengths determined from center-to-center distances as shown in Figure 3G and H. Uncertainties represent the standard deviation as discussed in Supplementary Methods 1. In OT experiments, only the DNA length bound to the tetrasome may be measured, as shown in Figures 4B and 5F (and only the value in Hmo1 contributes toward the average). Uncertainties are standard deviation for at least 10 arrays.
bThe number of nucleosomes per array is found in AFM experiments by identifying nucleosomes as shown in Supplementary Figure S1. Nucleosomes are only counted below a center-to-center distance of 75 nm (to exclude surface artifacts). In OT experiments, ripping events below ∼3 pN (Figures 4D and 5A) cannot be reliably counted so tetramers are assumed to be effectively dissociated from the DNA.
cThe HMGB binding affinity for the nucleosome is found from fits to AFM data and OT data. AFM data was fit to Supplementary Equation S5 as found in Supplementary Methods 4 and shown in Figure 3G and H. OT data was fit to Supplementary Equation S6 as found in Supplementary Methods 5 and shown in Figure 5B and C.
