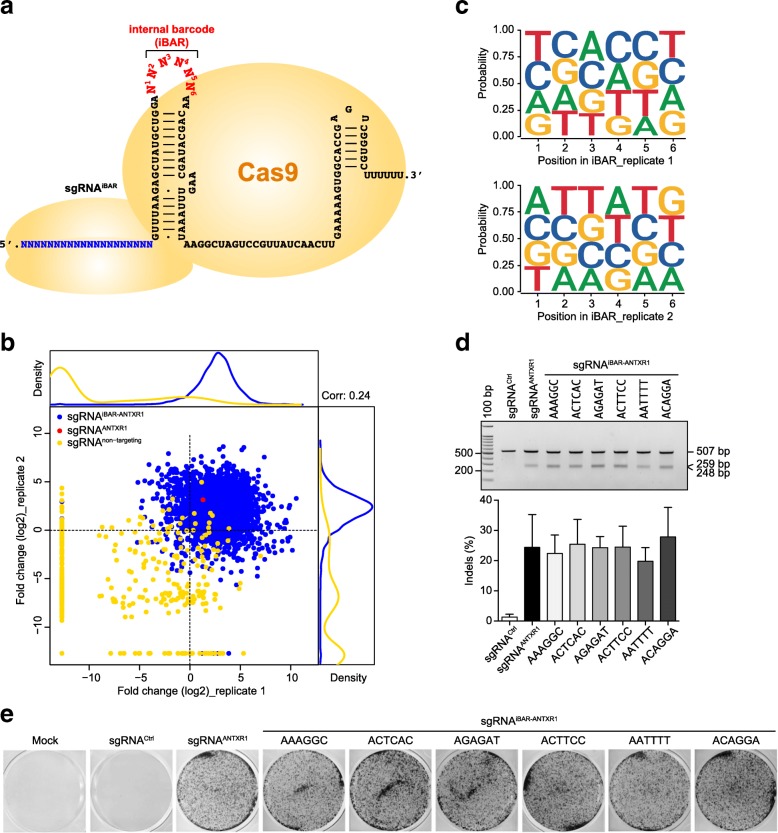
Fig. 1.
A design-based CRISPR screening via the gRNAiBAR method. a Schematic diagram of an sgRNA with an internal barcode (iBAR). A 6-nt barcode (iBAR6) was embedded in the tetra loop of the sgRNA scaffold. b CRISPR screening of a collection of one particular sgRNAANTXR1 containing all 4096 types of iBAR6. Fold changes between the reference and toxin treatment groups were calculated using the normalized abundances of sgRNAsANTXR1. A density plot of barcodes and non-targeting sgRNAs is presented. Corr: Pearson correlation. c Effects of nucleotides at each position of the iBAR on fold changes of guide RNAs. d Indels generated by sgRNAiBAR-ANTXR1 associated with six barcodes that appeared to be the worst in conferring cell resistance to PA/LFnDTA from the above screening. Percentages of cleavage efficiency in the T7E1 assay were measured using Image Lab software, and data are presented as the mean ± s.d. (n = 3). All primers used are listed in Additional file 3: Table S9. e MTT viability assay for the effects of the indicated sgRNAiBAR-ANTXR1 on the susceptibility of cells to PA/LFnDTA