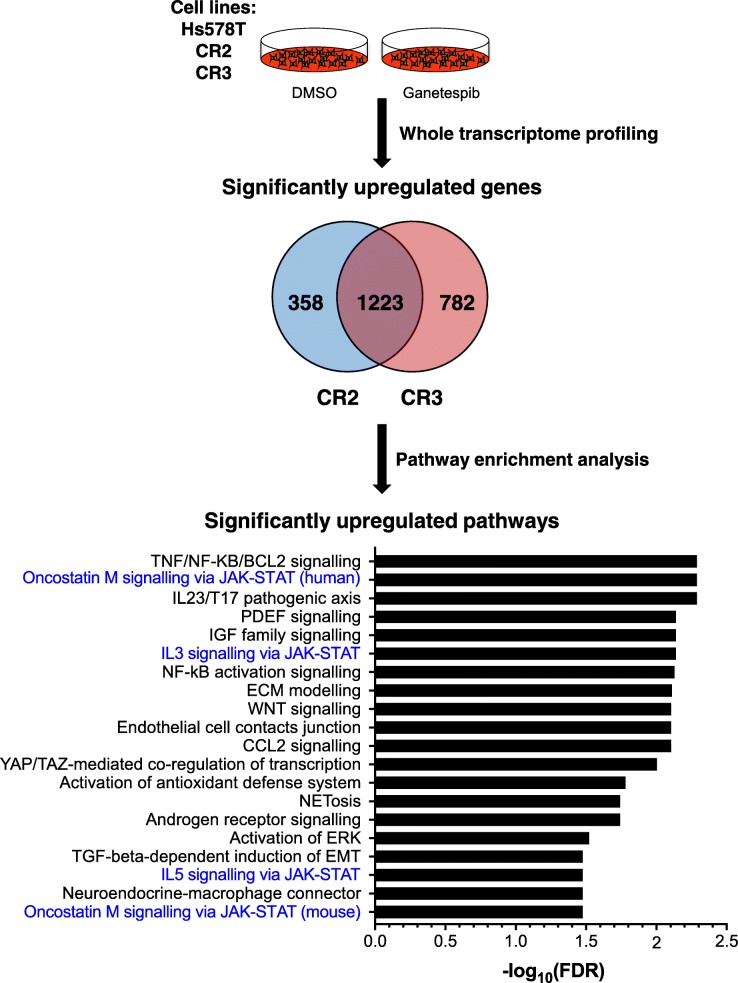
Fig. 3.
Differential RNA expression in HSP90i-resistant clones compared to parental Hs578T cells. RNA samples from DMSO-treated and ganetespib-treated Hs578T, CR2 and CR3 cells were analysed for whole transcriptome profiling with RNA-sequencing. Differential gene expression analyses were performed between DMSO-treated parental Hs578T cells with either DMSO-treated CR2 or CR3 and the significantly upregulated genes observed in these clones were mostly overlapping. Pathway enrichment analysis using Metacore™ was performed on the significantly upregulated overlapping genes. The graph represents the top 20 most significantly upregulated pathways in the HSP90i-resistant clones, with FDR (false discovery rate) value < 0.05. Pathways highlighted in blue are linked to JAK-STAT signalling