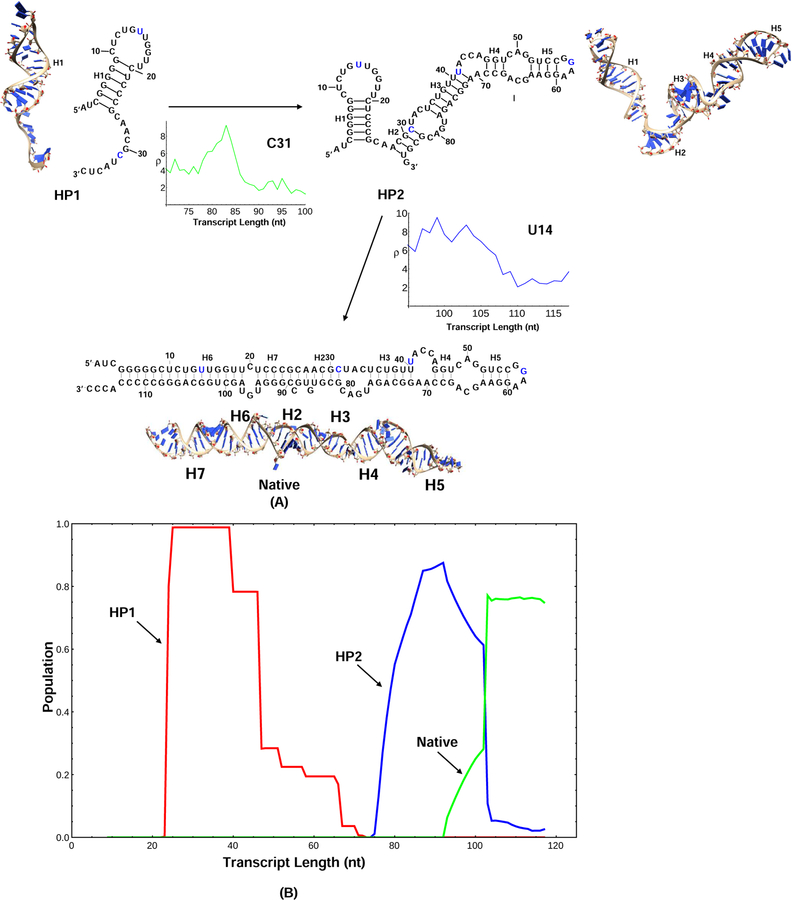
Figure 4:
(A) The figure shows that major SHAPE reactivity change for each transition, where the horizontal axis shows the elongating chain length during transcription and the vertical axis shows the SHAPE reactivity.27 During the transition from HP1 to HP2, C31 is paired with G82, the SHAPE reactivity of C31 begins to decrease after the chain is transcribed to the 83th nucleotide. From HP2 to the native structure, U14 become paired with G103 and the SHAPE profile also indicates the transition. The 3D structures for the kinetically important structures are predicted using the Vfold3D software55–58 and the Coarse-Grained MD simulations.59 (B) The population kinetics of 3 kinetically important conformations during the cotranscriptional folding process. The major transitions during the folding process are extracted from the predicted population kinetics for the different conformations. HP1 is first formed in the early transcription stage, followed by the formation of HP2, and additional helices and loops are added to HP1. In the late stage of the folding process, the first hairpin (H1) in HP2 is disrupted and the native structure is formed. From the population kinetics, the major folding pathway of E.Coli. SRP RNA is HP1→HP2→Native.