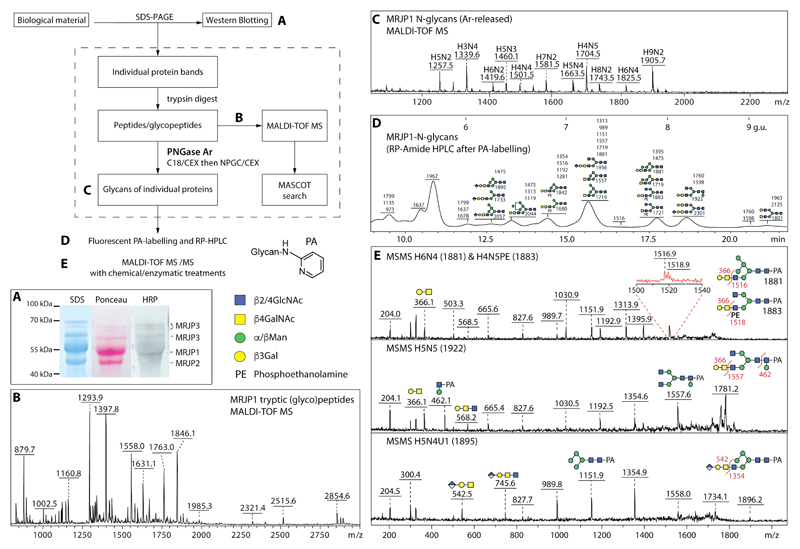
Figure 2. Example of a glycoproteomic study of honeybee glycoprotein MRJP1 found in royal jelly.
The flow chart scheme shows the glycoproteomic workflow for the biological sample; the letters refer to steps with exemplified by the following data. (A) N-glycan epitope detection by Western blotting of the royal jelly glycoproteins after incubation with anti-HRP antibodies. (B) Tryptic peptide mapping of one of the major royal jelly glycoproteins as measured with MALDI TOF MS in positive ion mode. (C) Free N-glycans from MRJP1 measured with MALDI-TOF MS after deglycosylation with PNGase Ar with the [M+Na]+ ions annotated with abbreviations of the form HxNy, where H is hexose and N is N-acetylhexosamine. (D) RP-HPLC chromatogram of reductively aminated N-glycans from royal jelly glycoprotein MRJP1 with fractions annotated with the detected glycan m/z values and calibrated in terms of glucose units. (E) MALDI-TOF MS/MS data of fractionated MRJP1 glycans, whereby [M+H]+ ions were fragmented and key B and Y ions are annotated. Note that the precursor ions for the H6N4 and H4N5PE structures cannot be separated (Δm/z = 2), but the zoom shows that Y ions derived from both are present; the structure of the PA (pyridylamino) reducing-terminal label as well as the symbol nomenclature are shown beside the flowchart.