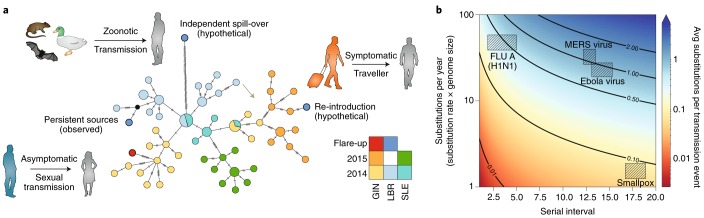
Fig. 2. Transmission chain tracking during outbreaks using virus genomics.
a, Viral genome sequences were used to distinguish between competing hypotheses for the source of the viruses that triggered the Ebola flare-ups in West Africa. The three main hypotheses and their expected genomic signatures are illustrated here with a hypothetical haplotype network. Genomes from all of the observed flare-ups grouped closely with genomes sequenced from patients in the same country, from earlier in the outbreak (bottom left), consistent with transmission from persistent sources. In contrast, genomes linked to re-introductions from neighbouring countries (right) would be expected to cluster with genomes from a different country and from late in the outbreak. In the case of independent spillovers from a reservoir host (top left, that is, independent sampling from the diversity circulating within the reservoir), the spillover genomes would be linked to the main outbreak by a long branch originating from near the root of the network. GIN, Guinea; LBR, Liberia; SLE, Sierra Leone. b, Expected ‘genomic resolution’ for the inference of transmission chains at the level of individual infections. Resolution is dependent on the serial interval between infections (x-axis; used as a proxy for epidemiological generation time), as well as the genome size and nucleotide substitution rate (y-axis). Icons courtesy of S. Knemeyer.