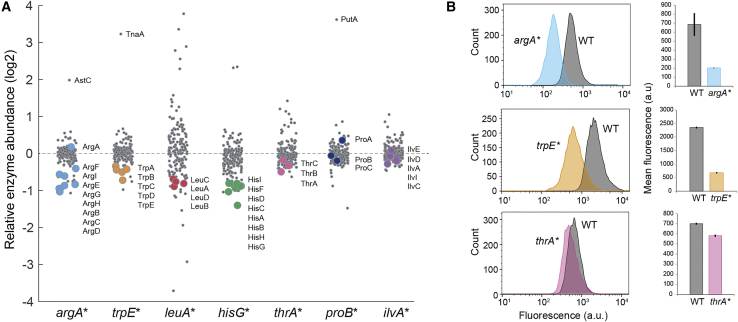
Figure 2.
Expression of Enzymes in Feedback-Dysregulated Pathways
(A) Abundance of 173 enzymes in amino acid metabolism (out of 204 enzymes in total), relative to the level in the wild-type. Data are represented as mean (n = 3). For each strain the enzymes in the dysregulated pathway are shown as colored dots. Enzymes in degradation pathways of arginine, tryptophan, and proline are indicated by their names.
(B) GFP-fluorescence measured by flow cytometry. GFP-promoter fusions were transformed in wild-type cells and the indicated mutant. Upper panel: pPargA-gfp; middle panel: pPtrpL-gfp; lower panel: pPthrL-gfp. Histograms represent fluorescence of 10,000 single cells. Mean fluorescence was calculated from 10,000 single cells of n = 3 independent cultures. See also Figure S3.