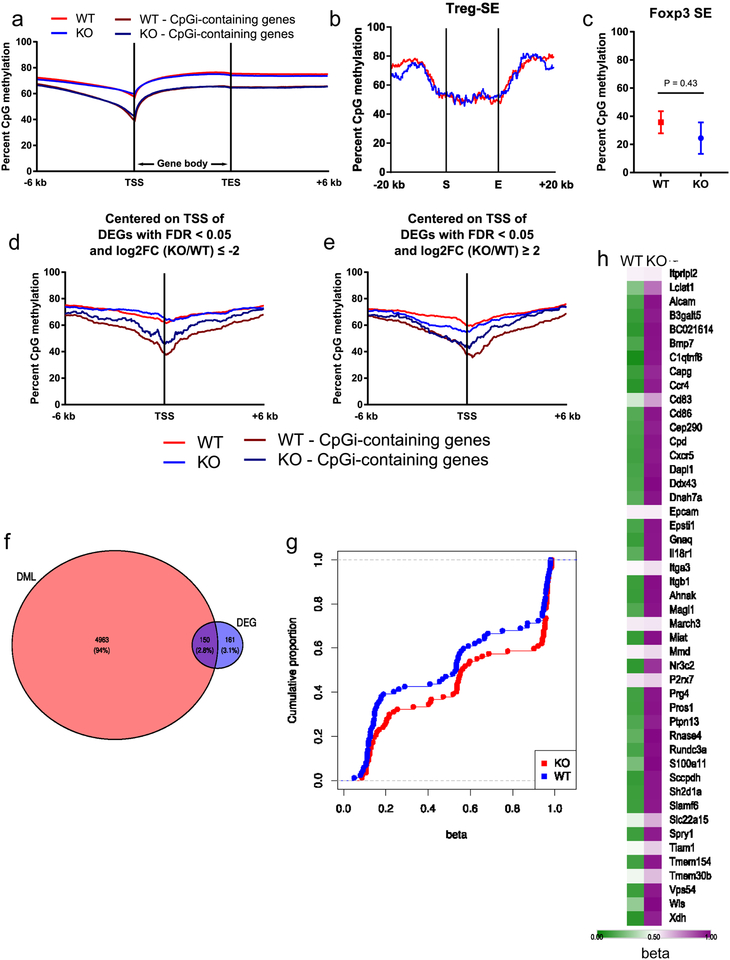
Extended Data Figure 8: Loss of RISP in Treg cells alters DNA methylation without affecting the Foxp3 locus in 3-week-old mice.
a, b, CpG methylation around the transcriptional start site (TSS) and transcriptional end site (TES) (a) and the start (S) and end (E) of Treg cell-specific super-enhancer elements with 20 kb of flanking sequence (b) in CD4+ Foxp3-YFP+ CD25+ cells from 3-week-old RISP KO (n=4) and RISP WT (n=4) animals. c, DNA methylation status of the super-enhancer associated with the Foxp3 locus in RISP KO and RISP WT Treg cells. d,e, Percentage of methylated CpGs around the TSS of differentially downregulated (d) and upregulated (e) genes (log2FC≥2, FDR q-value<.05) from Extended Data Figure 6. (a,d,e) mRRBS data were smoothed with sliding window size of 20 CpGs and a step of 10 CpGs for sites with >5X coverage. (b) Treg cell-specific super-enhancer data were normalized over 1-kb windows. Average CpG methylation of n=3 biological replicates per group is shown. f, Venn diagram analysis partitioning differentially methylated loci (DML) and differentially expressed genes (DEG). g, Cumulative distribution function of differentially methylated CpGs within 100 kilobases of 87 DEG that were down-regulated (log2 fold-change≤0) in chimeric KO Treg cells. Data points represent average methylation at each gene locus. h, Heatmap displaying the methylation state (beta) of differentially methylated CpGs at gene loci that were both hypermethylated and differentially down-regulated (adj-p<.01) in the chimeric KO Treg cells. (c) Data represent mean ± SEM and were analyzed with two-tailed t-test. (a-e) CD4+ Foxp3-YFP+ CD25+ cells from RISP WT (n=3) and RISP KO (n=3) animals were isolated on different days. Further processing and sequencing was performed with all 6 samples together. (f-h) CD4+ Foxp3-YFP+ CD25+ cells from RISP chimeric WT (n=4) and RISP chimeric KO (n=4) animals were isolated on different days. Further processing and sequencing was performed with all 8 samples together.