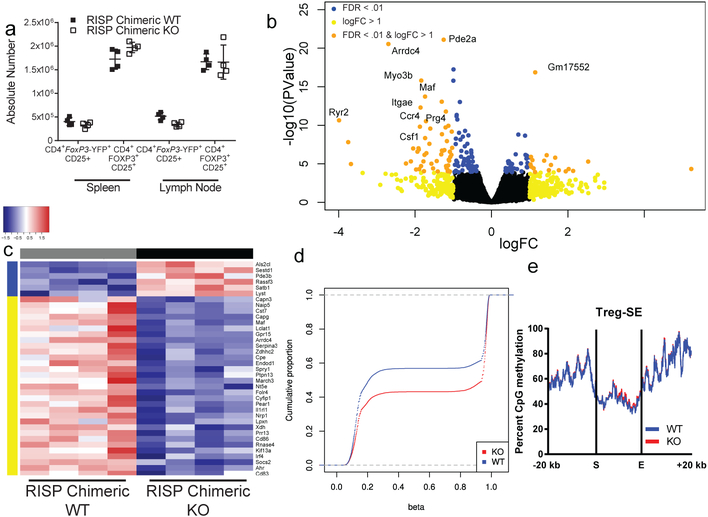
Figure 3: Complex III deficiency in Treg cells results in a cell autonomous impairment in expression of genes associated with Treg cell suppressive function along with DNA hypermethylation.
a, The absolute number of CD4+ Foxp3-YFP+ CD25+ (chimeric Treg cells) and CD4+ Foxp3+ CD25+ (total Treg cells) from adult RISP chimeric KO (n=4) and RISP chimeric WT (n=4) mice. b, Volcano plot showing differential gene expression in CD4+ Foxp3-YFP+ CD25+ cells isolated from adult RISP chimeric KO (n=4) and RISP chimeric WT (n=4) mice. c, Heat map of Treg cell signatures genes that are differentially expressed (adj. P<.01) in RISP chimeric KO (n=4) versus RISP chimeric WT (n=4) animals. d, Cumulative distribution function of 17,588 differentially methylated CpGs comparing chimeric Treg cells from RISP chimeric WT (n=4) and Treg-specific RISP-deficient chimeric KO (n=4) mice. Estimated CpG methylation is expressed as beta scores with 0 being unmethylated and 1 fully methylated. e, Metagene analysis of Treg cell-specific super-enhancer methylation from dataset in panel d. Data (a) represent mean ± SD and were analyzed with multiple two-tailed t-tests using a two-stage linear step-up procedure of Benjamini, Krieger and Yekutieli, with Q = 1%. Each cell type was analyzed individually, without assuming a consistent SD (Specific q-values displayed in supplemental source data). Data points on graphs represent individual animals isolated and analyzed on at least 2 separate days. Data in panels b-e are from one RNA sequencing (b,c) and one mRRBS (d,e) experiment respectively. CD4+ Foxp3-YFP+ CD25+ cells from RISP chimeric WT (n=4) and RISP chimeric KO (n=4) animals were isolated on different days. Further processing and sequencing was performed with all 8 samples together.