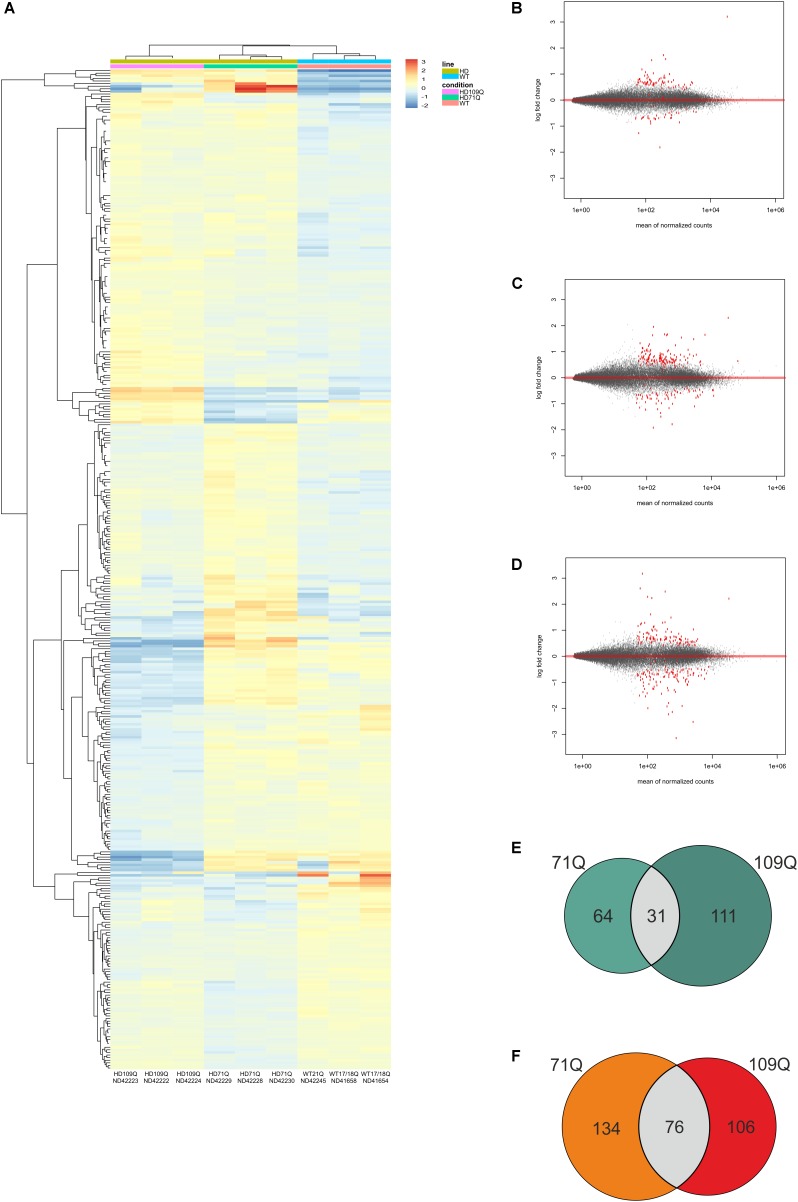
FIGURE 1.
RNA-seq Analysis of HD and control iPSC lines show early transcriptional changes in affected cells. (A) Heat map representing gene expression patterns of DE genes when comparing HD71Q and HD109Q iPSCs to control iPSCs at an adjusted p-value of < 0.05 and baseMean cutoff > 50. Red represents elevated expression while blue represent decreased expression, compared with the row mean. Each column represents each isogenic line. Gene names are shown on Supplementary Figure S3. (B–D) MA plots – differential expression analysis of significantly downregulated or upregulated mRNAs in HD iPSCs vs. control lines, as determined by population level RNA-seq – for HD vs. WT, 71Q vs. WT and 109Q vs. WT, respectively. (E) Venn diagram display of downregulated genes. (F) Venn diagram display of upregulated genes.