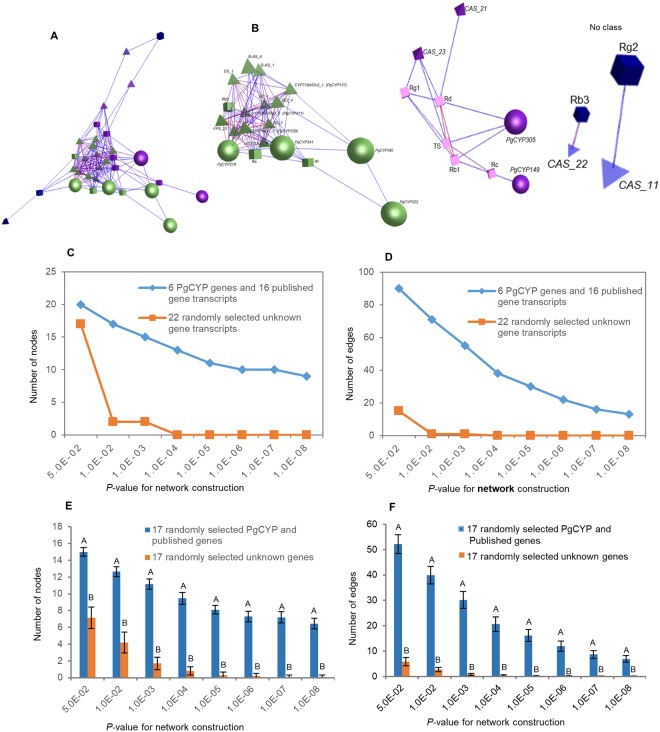
Figure 3.
Network analysis of the six ginsenoside biosynthesis candidate PgCYP genes and all 10 published ginsenoside biosynthesis genes. The network was constructed based on their expressions in the four-year-old roots of 42 cultivars. (A) The co-expression network constructed from the six PgCYP genes identified in this study indicated by balls, 16 transcripts of the 10 published ginsenoside biosynthesis genes indicated by diamonds, and 9 mono- and total ginsenosides indicated by cubes. The network was constructed at P ≤ 5.0E-02. (B) The clusters constituting the network. Different clusters are indicated by different colors. (C) Tendency that these ginsenoside biosynthesis related genes form a network, with the randomly-selected ginseng unknown genes as controls: variation in number of nodes. (D) Tendency that these ginsenoside biosynthesis related genes form a network, with the randomly-selected ginseng unknown genes as controls: variation in number of edges. (E) Statistics of variation in number of nodes in the network. (F) Statistics of variation in number of edges in the network. Different capital letters, significant at P ≤ 0.01. Error bar, standard deviation for 20 replications.