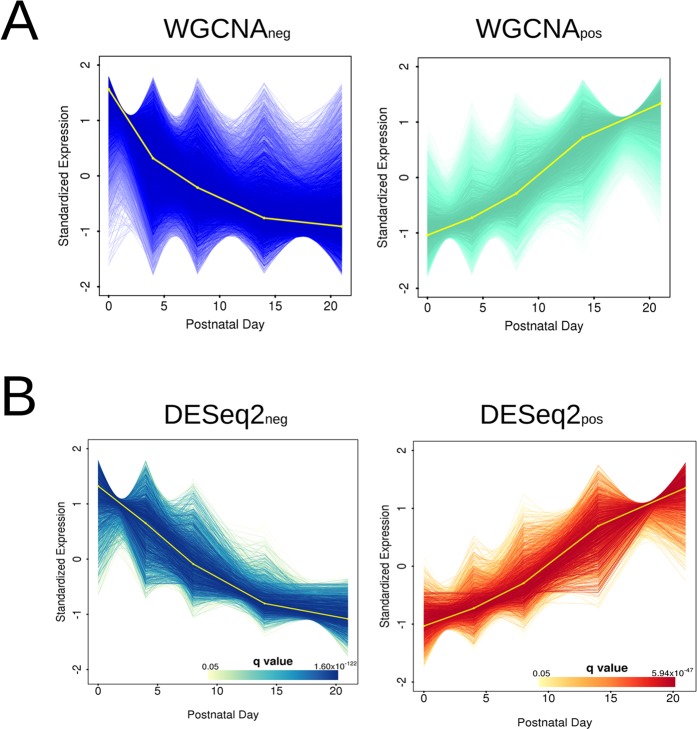
Figure 2.
Co-expression network analysis reveals temporal gene clusters that are correlated with PC development. (A) Genes assigned to the two WGCNA modules with statistically significant functional annotations (q < 0.05), expressing patterns of negative (WGCNAneg) and positive (WGCNApos) fold-change over time, respectively. WGCNAneg is significantly enriched for ontology terms associated with RNA processing, while WGCNApos is significantly enriched for ontology terms associated with neuron formation and neurological disorders. See also Supplementary Figs S2 and S3. Expression values are normalized and standardized to a mean of zero and standard deviation of 1. The eigengene of the cluster is displayed in yellow. (B) Statistically significant differentially expressed genes over time, separated in clusters of negative (DESeq 2neg) and positive (DESeq 2pos) fold-change, respectively. The DESeq 2 clusters show strong intersection with those identified by WGCNA and provide robustness for downstream analyses. Both WGCNA and DESeq 2 clusters contained similarly enriched ontology terms. See also Supplementary Fige S4 and Supplementary Table S3. Each gene within the module is plotted as a line of increasing colour intensity with decreasing q value, as displayed in the colour scale. Expression values are normalized and standardized to a mean of zero and standard deviation of 1. The eigengene of the cluster is displayed in yellow.