Abstract
We found an unusual Escherichia coli strain with resistance to colistin, carbapenem and amikacin from sewage. We therefore characterized the strain and determined the co-transfer of the resistance determinants. Whole genome sequencing was performed using both Illumina HiSeq X10 and MinION sequencers. Short and long reads were subjected to de novo hybrid assembly. Sequence type, antimicrobial resistance genes and plasmid replicons were identified from the genome sequences. Phylogenetic analysis of all IncHI2 plasmids carrying mcr-1 available in GenBank was performed based on core genes. Conjugation experiments were performed. mcr-3.5 was cloned into E. coli DH5α. The strain belonged to ST410, a type with a global distribution. Two colistin-resistant genes, mcr-1.1 and mcr-3.5, a carbapenemase gene blaNDM-5, and a 16S methylase gene rmtB were identified on different plasmids of IncHI2(ST3)/IncN, IncP, IncX3 and IncFII, respectively. All of the four plasmids were self-transmissible and mcr-1.1, mcr-3.5, blaNDM-5 and rmtB were transferred together. mcr-1-carrying IncHI2 plasmids belonged to several sequence types with ST3 and ST4 being predominant. MIC of colistin (4 μg/ml) for DH5α containing mcr-3.5 was identical to that containing the original mcr-3 variant. In conclusion, carbapenem resistance, colistin resistance and high-level aminoglycoside resistance can be transferred together even when their encoding genes are not located on the same plasmid. The co-transfer of multiple clinically-important antimicrobial resistance represents a particular challenge for clinical treatment and infection control in healthcare settings. Isolates with resistance to both carbapenem and colistin are not restricted to a given sequence type but rather are diverse in clonal background, which warrants further surveillance. The amino acid substitutions of MCR-3.5 have not altered its activity against colistin.
Introduction
Colistin is the last resort antimicrobial agent to treat infections caused by most Gram-negative bacteria commonly seen in clinical settings, including Escherichia coli, Klebsiella pneumoniae, Acinetobacter baumannii and Pseudomonas aeruginosa1,2. Bacterial strains that have acquired resistance to colistin have emerged worldwide. In addition to mutations or interruptions in certain chromosomal genes, acquired resistance to colistin has occurred due to plasmid-borne genes1. Eight plasmid-borne colistin resistance genes, i.e. mcr-13, mcr-24, mcr-35, mcr-46, mcr-57, mcr-68, mcr-79 and mcr-810, have been reported. The co-existence of two plasmid-borne colistin-resistant genes in bacterial isolates is uncommon, but recently, we reported the co-existence of mcr-1 and mcr-3 plus the carbapenemase gene blaNDM-5 in an E. coli clinical strain, WCHEC020123, of phylogenetic group A and sequence type 206 (ST206)11. Here we report a second independent occurrence of the co-existence of mcr-1, mcr-3 and blaNDM-5 in another E. coli strain, which was recovered from hospital sewage, belonging to a different sequence type (ST410). This strain has an even broader antimicrobial resistance spectrum than the extensive drug resistant WCHEC020123.
Materials and Methods
Recovery of the strain and in vitro antimicrobial susceptibility testing
E. coli strain WCHEC025943 was recovered from the influx mainstream of hospital sewage at West China Hospital, Chengdu, western China, in April 2017. The sewage sample was mixed with 100 ml brain heart infusion broth (Oxoid, Hampshire, UK) in a 500 ml flask. After overnight incubation at 37 °C, the culture suspension was diluted to 0.5 McFarland standard and an 100 μl aliquot was plated onto a CHROMAgar Orientation agar plate (CHROMAgar, Paris, France) containing 4 μg/ml colistin and 16 μg/ml meropenem. The plate was then incubated at 37 °C overnight. The pink colony that represents E. coli was screened for mcr-1 as described previously3. Species identification was established by Vitek II (bioMérieux, Marcy-l′Étoile, France) and by MALDI-TOF MS (Bruker, Billerica, MA, USA).
MICs of amikacin, aztreonam, aztreonam-avibactam, ceftazidime, ceftazidime-avibactam, ciprofloxacin, colistin, imipenem, meropenem, tigecycline and trimethoprim-sulfamethoxazole were determined using the broth microdilution method of the Clinical and Laboratory Standards Institute (CLSI)12. For ceftazidime-avibactam, colistin and tigecycline, the breakpoints defined by the European Committee on Antimicrobial Susceptibility Testing (EUCAST) (http://www.eucast.org/) were used, while the breakpoints of aztreonam were applied for aztreonam-avibactam.
Whole genome sequencing and analysis
Genomic DNA of strain WCHEC025943 was prepared using the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) and was subjected to whole genome sequencing using both the HiSeq X10 platform (Illumina, San Diego, CA, USA) and the long-read MinION Sequencer (Nanopore, Oxford, UK). The de novo hybrid assembly of both short Illumina reads and long MinION reads was performed using Unicycler13 under conservative mode for increased accuracy. Complete circular contigs generated were then corrected using Plion14 with Illumina reads for several rounds until no change was detected.
Sequence type was determined using the genome sequence to query the E. coli multi-locus sequence typing database (http://enterobase.warwick.ac.uk/species/index/ecoli). Antimicrobial resistance genes were identified from genome sequences using ResFinder at https://cge.cbs.dtu.dk/services/ResFinder/. Plasmid replicon types and sequence types of IncHI2 and IncF plasmids were determined using PlasmidFinder and pMLST tools at https://cge.cbs.dtu.dk/services/PlasmidFinder/ and https://cge.cbs.dtu.dk/services/pMLST/. Single nucleotide polymorphisms (SNPs) between strain WCHEC025943 and strain WCHEC14828 (also called WCHEC005828, GenBank accession no. RIAW00000000), a blaOXA-181-carrying ST410 E. coli identified in the same hospital in 201415, was determined from a two-way whole genome alignment in HarvestTools16.
Nucleotide sequence accession numbers
Complete sequences of the chromosome and plasmids of strain WCHEC025943 have been deposited into GenBank under the accession no. CP027199 to CP027205.
Phylogenetic group typing
E. coli phylogenetic group of strain WCHEC025943 was determined using PCR as described previously17.
Cloning of mcr-3.5
The complete coding sequence of mcr-3.5 was amplified with primers mcr3.5-up (CTGGTCGGAGATATGGGTGT) and mcr3.5-dw (GGCATTCAACATCAGAGCAA) using PrimeSTAR Max DNA Polymerase (Takara, Dalian, China). The primers were designed to amplify the gene with 222-bp upstream and 540-bp downstream sequences of mcr-3.5. Amplicons were ligated to the pMD20-T vector using the Mighty TA-cloning kit (Takara). The ligated fragments were transformed into E. coli DH5α. mcr-3.5-containing transformants were selected on LB agar plates containing 2 μg/mL colistin. The presence of mcr-3.5 in transformants was confirmed by PCR. MIC of colistin was determined for transformants carrying mcr-3.5 using the broth microdilution method12.
Phylogenetic analysis of IncHI2 plasmids
Complete sequences of all IncHI2 plasmids carrying mcr-1 (n = 25 in addition to pMCR1_025943 and pMCR1_020123) were retrieved from GenBank. Plasmid replicon types and sequence types of these plasmids were determined using PlasmidFinder and pMLST. Annotation was performed using Prokka18 and antimicrobial resistance genes were identified using ResFinder. Orthologues of these plasmids were identified using OrthoFinder19 with default settings, resulting in a sum of 56 genes representing the core genome of these 27 plasmids. The alleles of orthologous genes were aligned using MAFFT20 and concatenated into a single sequence containing 56 aligned genes for each plasmid. The maximum-likelihood phylogenetic tree was inferred based on the core genome using RAxML21 with a 1000-bootstrap test.
Conjugation
Conjugation experiments were carried out in brain heart infusion broth at 30 °C using azide-resistant E. coli strain J53 as the recipient. Transconjugants were selected on LB agar plates containing 150 μg/ml sodium azide plus 2 μg/ml colistin for mcr-1.1 and mcr-3.5, plus 1 μg/ml meropenem for blaNDM-5 or plus 64 μg/ml amikacin for rmtB. Transconjugants were also selected on LB agar plates containing 150 μg/ml sodium azide plus 2 μg/ml colistin, 1 μg/ml meropenem and 64 μg/ml amikacin to examine whether mcr, blaNDM-5 and rmtB could be transferred together. The presence of mcr-1.1, mcr-3.5, blaNDM-5 and/or rmtB in transconjugants was screened using PCR and Sanger sequencing. Conjugation frequency was calculated as the number of transconjugants per recipient cell.
Results and Discussion
Strain WCHEC025943 was recovered from the sewage sample and grew on the agar plate containing 4 μg/ml colistin and 16 μg/ml meropenem. The complete genome sequence of strain WCHEC025943 was obtained, which was 5.1 Mb and contained a 4.82 Mb circular chromosome and six plasmids of different replicon types (Table 1).
Table 1.
Plasmids and antimicrobial resistance genes in strain WCHEC0259431.
| Plasmid | Replicon type | Size (bp) | Antimicrobial resistance genes | ||||
|---|---|---|---|---|---|---|---|
| Colistin resistance | Carbapenemase | ESBL | 16S methylase | Others | |||
| p1_025943 | Y | 95,859 | — | ||||
| p2_025943 | FII, FIB | 75,779 | rmtB | aac(3)-IId, aac(6′)-Ib-cr, aadA16, aph(3″)-Ib, aph(6)-Id, arr-3, blaTEM-1B, dfrA14, dfrA27, sul1, sul2, tet(A) - | |||
| p3_025943 | Col (BS512) | 2,088 | — | ||||
| pMCR1_025943 | HI2, N | 265,538 | mcr-1.1 | bla CTX-M-65 | aac(3)-IVa, aadA22, aph(3′)-Ia, aph(3″)-Ib, aph(6)-Id, aph(4)-Ia, blaTEM-1B, floR, lnu(F), strA, mphA, oqxA, oqxB, sul1, sul2, tet(A), tet(M) | ||
| pMCR3_025943 | P | 50,520 | mcr-3.5 | — | |||
| pNDM5_025943 | X3 | 45,275 | bla NDM-5 | — | |||
1blaCMY-2 was located on the chromosome.
The strain was resistant to amikacin (>512 μg/ml), aztreonam (>512 μg/ml), ceftazidime (>512 μg/ml), ceftazidime-avibactam (>512/4 μg/ml), ciprofloxacin (8 μg/ml), colistin (8 μg/ml), imipenem (128 μg/ml), meropenem (128 μg/ml) and trimethoprim-sulfamethoxazole (128/2432 μg/ml), but was susceptible to aztreonam-avibactam (1/4 μg/ml) and tigecycline (0.5 μg/ml). Strain WCHEC025943 had 31 known acquired antimicrobial resistance genes mediating resistance to aminoglycosides (aac(3)-IVa, aac(3)-IId, aac(6′)-Ib-cr, aadA22, aadA16, aph(3′)-Ia, aph(3″)-Ib, aph(6)-Id, aph(4)-Ia, rmtB, strA), β-lactams (blaCTX-M-65, blaCMY-2, blaNDM-5, blaTEM-1B), fosfomycin (fosA), colistin (mcr-1.1, mcr-3.5), macrolide-lincosamide-streptogramin B (lnu(F), mphA), phenicol (floR), rifampicin (arr-3), quinolones (aac(6′)-Ib-cr, oqxA, oqxB), sulphonamides (sul1, sul2), tetracycline (tet(A), tet(M)) and trimethoprim (dfrA14, dfrA27). Compared with amikacin-susceptible strain WCHEC020123 carrying mcr-1.1, mcr-3.5 and blaNDM-511, strain WCHEC025943 had rmtB, which could explain its high-level resistance to amikacin. As a metallo-β-lactamase-encoding gene, blaNDM-5 did not confer resistance to aztreonam but the presence of blaCTX-M-65 (encoding an extended-spectrum β-lactamase) and blaCMY-2 (encoding an AmpC cephalosporinase) completed the resistance to all commercially available β-lactams including aztreonam in strain WCHEC025943.
Of note, mcr-3.5 encodes three amino acid substitutions (M23V, A456E and T488I) compared with the original mcr-3 variant on plasmid pWJ1 (GenBank accession no. KY924928). MCR-3 has been predicted to have two domains, i.e. Domain 1 (residues 1 to 172) containing 5 transmembrane α-helices and Domain 2 (residues 173 to 541), a periplasmic domain containing the putative catalytic center5. The amino acid substitutions of MCR-3.5 occurred in the first α-helix (M23V) of Domain 1 and in Domain 2 (A456E and T488I). However, the MIC of colistin for E. coli DH5α containing mcr-3.5 was 4 μg/ml, which was identical to that for E. coli containing the original mcr-3 variant5. This confirms that the amino acid substitutions of MCR-3.5 have not altered its activity against colistin as described previously11.
Unlike the ST206 strain WCHEC020123, strain WCHEC025943 belonged to ST410 and phylogenetic group A. ST410 (adk-fumC-gyrB-icd-mdh-purA-recA, 6-4-12-1-20-18-7) and ST206 (6-7-5-1-8-18-2) shared only three out of the seven alleles for MLST. ST410 E. coli has a worldwide distribution22–24 and a blaOXA-181-carrying ST410 E. coli, strain WCHEC14828, has been identified in the same hospital in 201415. There were 109 SNPs between strain WCHEC025943 and WCHEC14828, suggesting divergence from a relatively recent common ancestor and differential plasmid acquisition and maintenance. The co-existence of mcr-1 and blaNDM-5 has been found in E. coli isolates of various STs, such as ST156, ST446 and ST64825,26. Strains with resistance to both carbapenem and colistin are therefore not restricted to distinct lineages but rather are diverse in clonal background.
The rmtB gene in strain WCHEC025943 was carried on a 75.8-kb IncF plasmid containing an IncFII replicon (FII_47 allele) and an IncFIB replicon (a new allele). By contrast, strain WCHEC020123 did not have IncF plasmids. Like strain WCHEC020123, in strain WCHEC025943, mcr-1.1, mcr-3.5 and blaNDM-5 were carried by three plasmids belonging to different replicon types (Table 1). blaNDM-5 was carried by an IncX3 plasmid, which was almost identical to the blaNDM-5-carrying IncX3 plasmid in strain WCHEC020123. mcr-3.5 was carried on a 50.5-kb IncP plasmid, designated pMCR3_025943, in strain WCHEC025943. pMCR3_025943 is identical to pMCR3_020123, the mcr-3.5-carrying IncP plasmid in strain WCHEC02012311, except that an insertion sequence, IS1294, is absent from pMCR3_025943 but is inserted in a spacer region in pMCR3_020123. mcr-1 was carried on a 265.5-kb plasmid (designated pMCR1_025943) containing both IncHI2 (ST3) and IncN replicons in strain WCHEC025943, which was larger than the 223.7-kb mcr-1-carrying IncHI2 (ST3) plasmid (pMCR1_020123) in strain WCHEC020123. The major differences between the two ST3-IncHI2 plasmids, pMCR1_025943 and pMCR1_020123, are the presence of IncN replicon and a 30-kb region containing several genes (traN, traU, traW) encoding conjugation in the former but absent from the latter. ST3-IncHI2 plasmids have been found increasingly as the vector of mcr-1 and are particularly large and complex in structure with the ability to acquire multiple antimicrobial resistance genes and additional plasmid replicons27–29.
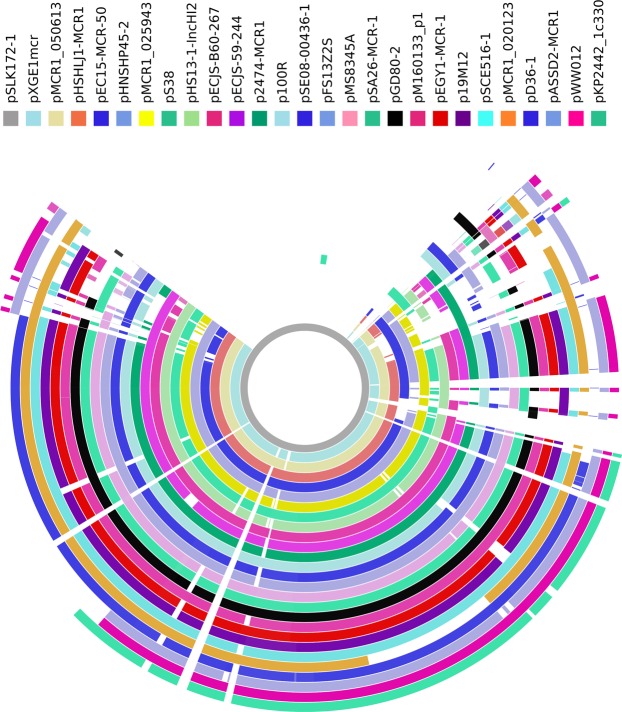
mcr-1-carrying IncHI2 plasmids were mostly found in E. coli and were also present in several other species of the Enterobacteriaceae (Fig. 1). A few IncHI2 plasmids also contain additional replicons, among which IncN replicon was the most common (Fig. 1). These plasmids were large in size (125,572 to 256,620 bp for plasmids containing IncHI2 replicons alone and 238,539 to 369,298 bp for those containing additional replicons) and commonly carried multiple antimicrobial resistance genes (Fig. 1). Most of these plasmids belong to ST3 (n = 15) or ST4 (n = 8), while one belongs to ST14 and the sequence type is not assigned to three plasmids due to the absence of an allele for IncHI2 pMLST. This suggests that several types of IncHI2 plasmids could mediate the transfer of mcr-1 and ST3-IncHI2 is the most common type (Fig. 1). These plasmids were also aligned against pSLK172-1 (GenBank accession no. CP017632), the largest (369,298 bp) mcr-1-carrying IncHI2 plasmid, using BRIG30. This revealed that mcr-1-carrying IncHI2 plasmids are complex and highly variable in structure (Fig. 2).
Figure 1.
Phylogenetic analysis of mcr-1-carrying IncHI2 plasmids. Plasmid name, sequence type, replicon other than IncHI2, host species, plasmid size, GenBank accession no. and antimicrobial resistance genes carried are shown. Bootstrap values are indicated by dots with different degrees of colour.
Figure 2.
Alignment of mcr-1-carrying IncHI2 plasmids. Alignment was performed using BRIG30. pSLK172-1 (GenBank accession no. CP017632) was used as the reference due to the fact that it is the largest (369,298 bp) mcr-1-carrying IncHI2 plasmid and contains additional IncN and IncY replicons. GenBank accession no. of these plasmids are shown in Fig. 1.
In strain WCHEC025943, the four plasmids carrying mcr-1.1, mcr-3.5, blaNDM-5 or rmtB were all self-transmissible at a 10−3, 10−4, 10−3 and 10−4 frequency, respectively. Alarmingly, the four plasmids could be transferred together to a single transconjugant at a 10−6 frequency. This suggests that carbapenem resistance, colistin resistance and aminoglycoside resistance can be transferred together even when their encoding genes are located on separate plasmids.
Conclusion
The above findings suggest that carbapenem resistance, colistin resistance and high-level aminoglycoside resistance can be transferred together even when their encoding genes are not located on the same plasmid. The co-transfer of multiple clinically-important antimicrobial resistance represents a particular challenge for clinical treatment and infection control in healthcare settings, which warrant more surveillance and further studies to explore counter measures. Isolates with resistance to both carbapenem and colistin are not restricted to a given sequence type but rather are diverse in clonal background. mcr-1-carrying IncHI2 plasmids belonged to several sequence types with ST3 and ST4 being predominant.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (Project Nos 81772233, 81661130159 and 81572030) and the Newton Advanced Fellowship, Royal Society, UK (NA150363).
Author Contributions
Z.Z. designed the study. H.L., Y.F., K.M. and L.L. collected the data. H.L., A.M. and Z.Z. analyzed and interpreted the data. Z.Z. wrote the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Poirel L, Jayol A, Nordmann P. Polymyxins: antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin Microbiol Rev. 2017;30:557–596. doi: 10.1128/CMR.00064-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Olaitan AO, Morand S, Rolain JM. Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front Microbiol. 2014;5:643. doi: 10.3389/fmicb.2014.00643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu Y-Y, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 4.Xavier BB, et al. Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia coli, Belgium, June 2016. Euro Surveill. 2016;21:30280. doi: 10.2807/1560-7917.ES.2016.21.27.30280. [DOI] [PubMed] [Google Scholar]
- 5.Yin W, et al. Novel plasmid-mediated colistin resistance gene mcr-3 in Escherichia coli. mBio. 2017;8:e00543–00517. doi: 10.1128/mBio.00543-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carattoli A, et al. Novel plasmid-mediated colistin resistance mcr-4 gene in Salmonella and Escherichia coli, Italy 2013, Spain and Belgium, 2015 to 2016. Euro Surveill. 2017;22:30589. doi: 10.2807/1560-7917.ES.2017.22.31.30589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borowiak M, et al. Identification of a novel transposon-associated phosphoethanolamine transferase gene, mcr-5, conferring colistin resistance in D-tartrate fermenting Salmonella enterica subsp. enterica serovar Paratyphi B. J Antimicrob Chemother. 2017;72:3317–3324. doi: 10.1093/jac/dkx327. [DOI] [PubMed] [Google Scholar]
- 8.AbuOun M, et al. mcr-1 and mcr-2 variant genes identified in Moraxella species isolated from pigs in Great Britain from 2014 to 2015. J Antimicrob Chemother. 2017;72:2745–2749. doi: 10.1093/jac/dkx286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang YQ, Li YX, Lei CW, Zhang AY, Wang HN. Novel plasmid-mediated colistin resistance gene mcr-7.1 in Klebsiella pneumoniae. J Antimicrob Chemother. 2018;73:1791–1795. doi: 10.1093/jac/dky111. [DOI] [PubMed] [Google Scholar]
- 10.Wang X, et al. Emergence of a novel mobile colistin resistance gene, mcr-8, in NDM-producing Klebsiella pneumoniae. Emerg Microbes Infect. 2018;7:122. doi: 10.1038/s41426-018-0124-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu L, Feng Y, Zhang X, McNally A, Zong Z. New variant of mcr-3 in an extensively drug-resistant Escherichia coli clinical isolate carrying mcr-1 and blaNDM-5. Antimicrob Agents Chemother. 2017;61:01757–01717. doi: 10.1128/AAC.01757-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.CLSI. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Seventh Informational Supplement. M100-S27. (Clinical and Laboratory Standards Institute 2017).
- 13.Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol. 2017;13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walker BJ, et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One. 2014;9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Y, et al. First report of OXA-181-producing Escherichia coli in China and characterization of the isolate using whole-genome sequencing. Antimicrobial Agents and Chemotherapy. 2015;59:5022–5025. doi: 10.1128/AAC.00442-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Treangen TJ, Ondov BD, Koren S, Phillippy AM. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 2014;15:524. doi: 10.1186/s13059-014-0524-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clermont O, Christenson JK, Denamur E, Gordon DM. The Clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environ Microbiol Rep. 2013;5:58–65. doi: 10.1111/1758-2229.12019. [DOI] [PubMed] [Google Scholar]
- 18.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 19.Emms DM, Kelly S. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 2015;16:157. doi: 10.1186/s13059-015-0721-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vervoort J, et al. High rates of intestinal colonisation with fluoroquinolone-resistant ESBL-harbouring Enterobacteriaceae in hospitalised patients with antibiotic-associated diarrhoea. Eur J Clin Microbiol Infect Dis. 2014;33:2215–2221. doi: 10.1007/s10096-014-2193-9. [DOI] [PubMed] [Google Scholar]
- 23.Chen YT, et al. KPC-2-encoding plasmids from Escherichia coli and Klebsiella pneumoniae in Taiwan. J Antimicrob Chemother. 2014;69:628–631. doi: 10.1093/jac/dkt409. [DOI] [PubMed] [Google Scholar]
- 24.Huber H, Zweifel C, Wittenbrink MM, Stephan R. ESBL-producing uropathogenic Escherichia coli isolated from dogs and cats in Switzerland. Vet Microbiol. 2013;162:992–996. doi: 10.1016/j.vetmic.2012.10.029. [DOI] [PubMed] [Google Scholar]
- 25.He T, et al. Characterization of NDM-5-positive extensively resistant Escherichia coli isolates from dairy cows. Vet Microbiol. 2017;207:153–158. doi: 10.1016/j.vetmic.2017.06.010. [DOI] [PubMed] [Google Scholar]
- 26.Yang RS, et al. Emergence of NDM-5- and MCR-1-producing Escherichia coli clones ST648 and ST156 from a single muscovy duck (Cairina moschata) Antimicrob Agents Chemother. 2016;60:6899–6902. doi: 10.1128/AAC.01365-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cain AK, Hall RM. Evolution of IncHI2 plasmids via acquisition of transposons carrying antibiotic resistance determinants. J Antimicrob Chemother. 2012;67:1121–1127. doi: 10.1093/jac/dks004. [DOI] [PubMed] [Google Scholar]
- 28.Fang LX, et al. High genetic plasticity in multidrug resistant ST3-IncHI2 plasmids revealed by sequence comparison and phylogenetic analysis. Antimicrob Agents Chemother. 2018;62:e02068–02017. doi: 10.1128/AAC.02068-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao F, Feng Y, Lu X, McNally A, Zong Z. Remarkable diversity of Escherichia coli carrying mcr-1 from hospital sewage with the identification of two new mcr-1 variants. Front Microbiol. 2017;8:2094. doi: 10.3389/fmicb.2017.02094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics. 2011;12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]