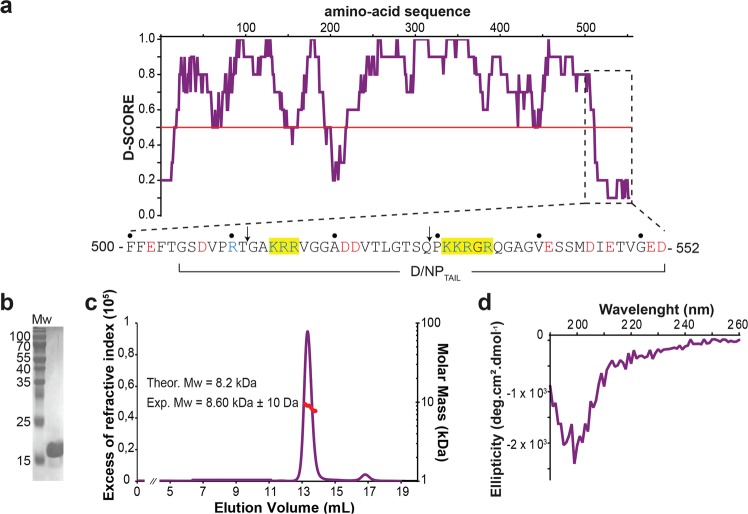
Figure 4.
Biophysical characterization of D/NPTAIL. (a) D-score (score for disorder as a function of residue) of D/NP with a zoom (below the graph) on the last 50 residues. The prediction is based on 22 predictor web servers and the D-score was calculated by adding the values for each residue and dividing by the number of used algorithms. We arbitrarily defined a threshold level at 0.50; residues with a D-score <0.50 were assigned as disordered38. The yellow boxes on the sequence are to highlight the putative NLS motifs. The arrows indicate where the sequences were cut for making D/NP-529 and D/NP-511. (b) Coomassie blue-stained SDS-PAGE (Tris-Tricine; 15% polyacrylamide) of the purified D/NPTAIL. It migrates at a higher molecular weight (17 kDa approximately) than expected (8 kDa). (c) SEC-MALLS-RI analysis of D/NPTAIL loaded on S75 10/300 GL column. For this experiment, we have chosen to keep the His-tag encoded with the pETM11 plasmid, for an optimal detection of D/NPTAIL with UV. The experimental molecular weight is consistent with the expected mass. (d) Circular dichroïsm of D/NPTAIL. CD is a biophysical method based on the polarization of light, used for a fast determination of the secondary structures within the proteins in solution. α-Helical proteins show negative bands at 222 nm and 208 nm and a positive band at 193 nm, proteins with well-defined antiparallel β-sheets have negative bands at 218 nm and positive bands at 195 nm and disordered proteins have very low ellipticity above 210 nm and negative bands near 195 nm.