Figure 1.
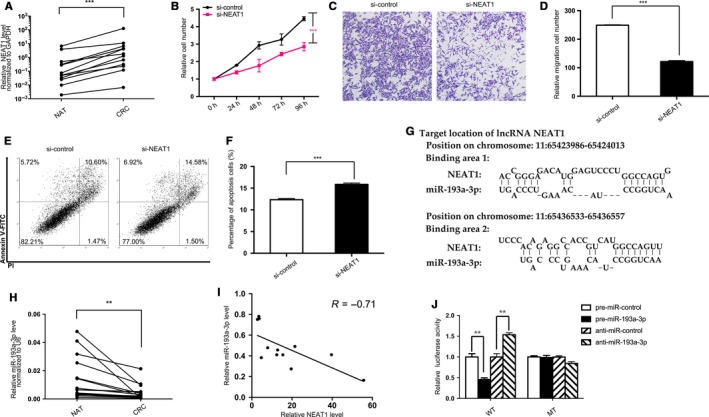
Prediction and confirmation of NEAT1/miR‐193a‐3p's binding. A, Quantitative RT‐PCR analysis of the expression levels (NEAT1 lncRNA vs GAPDH mRNA) of NEAT1 in the 12 pairs of colorectal cancer (CRC) and normal adjacent tissues (NAT). B‐F, Cell functional assays performed after the transfection of SW480 cells with equal doses of NEAT1 siRNA (si‐NEAT1) or scrambled negative control RNA (si‐control). B, Proliferation. C, Migration. D, The number of migrated cells was quantified. E, Apoptosis. F, The percentage of apoptosis cells was quantified. G, Predicted miR‐193a‐3p of seed sequences targeting NEAT1 by LncBase v2. H, Quantitative RT‐PCR analysis of the expression levels (miR‐193a‐3p vs U6) of miR‐193a‐3p in the aforementioned CRC and NAT samples. I, Pearson's correlation scatter plot of the fold change of NEAT1 and miR‐193a‐3p in CRC samples. J, Recognition of the NEAT1 seed sequences by miR‐193a‐3p. Firefly luciferase reporters containing either wild‐type (WT) or mutant (MUT) miR‐193a‐3p binding sites in NEAT1 were co‐transfected into 293T cells with equal doses of miR‐193a‐3p mimics, inhibitors, or scrambled negative control RNAs. Forty‐eight hours post‐transfection, the cells were assayed using a luciferase assay kit. The results are presented as the mean ± SD of three independent experiments. (**P < 0.01; ***P < 0.001).
