Figure 2.
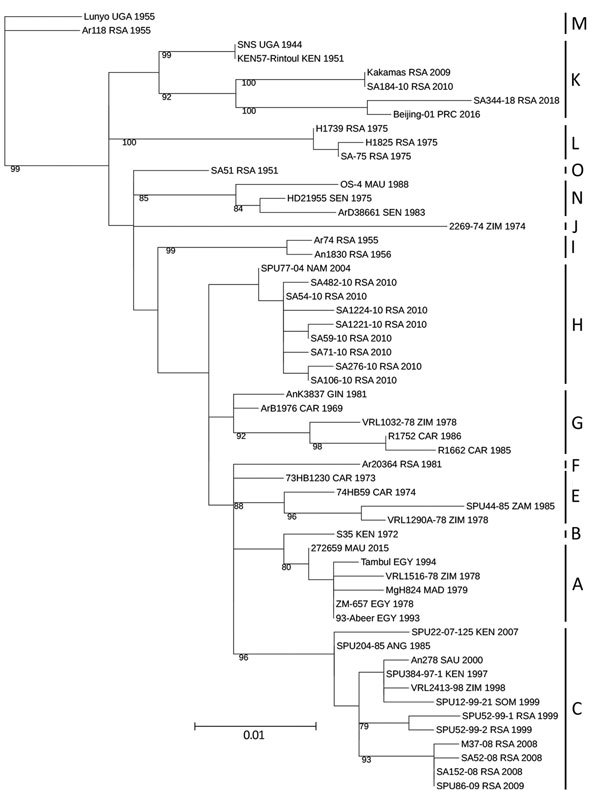
Maximum-likelihood tree showing the phylogeny of Rift Valley fever virus isolate SA344-18, collected in South Africa in May 2018, on the basis of a 490-nt fragment of the medium segment. Lineage names according to the nomenclature of Grobbelaar et al. (5) are indicated. Maximum-likelihood analysis was performed in RAxML version 8.2.10 (http://evomics.org/learning/phylogenetics/raxml); 100,000 bootstrap replicates were performed. Bootstrap values are shown at the nodes. Scale bar indicates nucleotide changes per base pair. ANG, Angola; CAR, Central African Republic; EGY, Egypt; GIN, Guinea; KEN, Kenya; MAD, Madagascar; MAU, Mauritania; NAM, Namibia; PRC, China; RSA, South Africa; SAU, Saudi Arabia; SEN, Senegal; SOM, Somalia; UGA, Uganda; ZAM, Zambia; ZIM, Zimbabwe.
