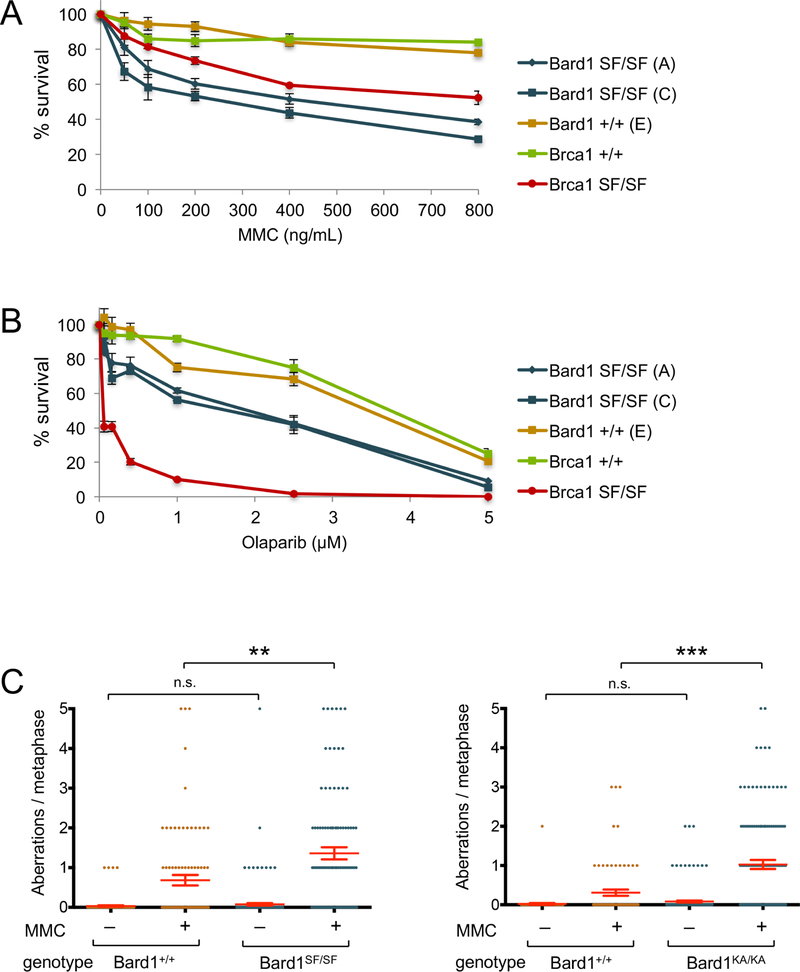
Figure 1. Bard1SF/SF cells are hypersensitive to DNA damaging agents and display genotoxin-induced chromosomal instability.
A) Colony survival analysis of MMC-treated isogenic Bard1+/+ and Bard1SF/SF MEFs, along with Brca1+/+ and Brca1SF/SF MEFs. Survival is quantified as percentage of colonies on MMC-treated relative to untreated plates. Each condition was tested in triplicate, and error bars represent standard error of the mean.
B) Colony survival analysis of olaarib-treated Bard1+/+ and Bard1SF/SF MEFs, along with Brca1+/+ and Brca1SF/SF MEFs.
C) Bard1+/+ and Bard1SF/SF primary MEFs were cultured with or without 40 ng/mL MMC for 16 hours and structural chromosome abnormalities were quantified by T-FISH. The mean number of aberrations per cell is denoted by a horizontal red line, and the error bars represent standard error of the mean. P values were calculated by unpaired Student’s T-Test (n.s. = no significance, ** = p<0.01, *** = p<0.001).
D) T-FISH analysis of Bard1+/+ and Bard1KA/KA primary MEFs.