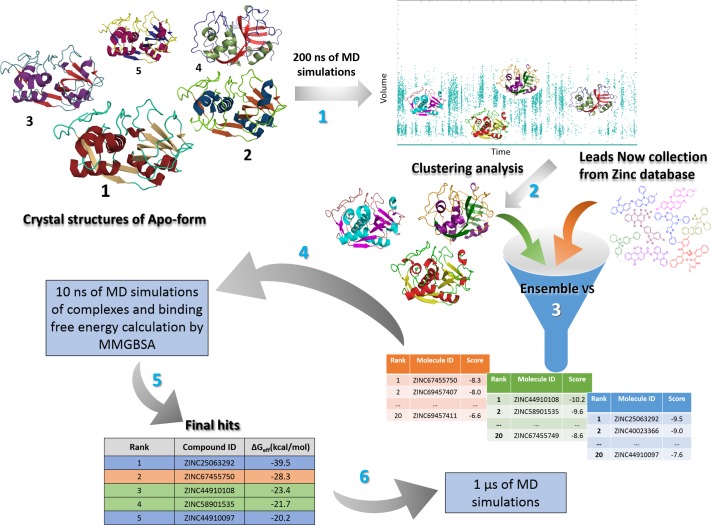
Fig 2. Workflow of structure-based identification of cruzain allosteric inhibitors.
In the first step, five cruzain apo structures were selected and subjected to 200 ns of MD simulations. The second step represents the volume-based cluster analysis performed for each allosteric site, which produced three representative structures per pocket. The third step illustrates the ensemble VS accomplished for each cavity, where twenty hits were chosen per representative structure (60 per allosteric site). Subsequently, 10 ns MD simulations were carried out with these 60 complexes and the respective trajectories were employed for MM-GBSA binding free energy calculations. In this step, a final list of five hit compounds, ranked according to the MM-GBSA results, was obtained per allosteric site. Finally, 200 ns MD simulations were conducted to analyze the potentially mechanism of cruzain allosteric modulation exerted by these compounds.