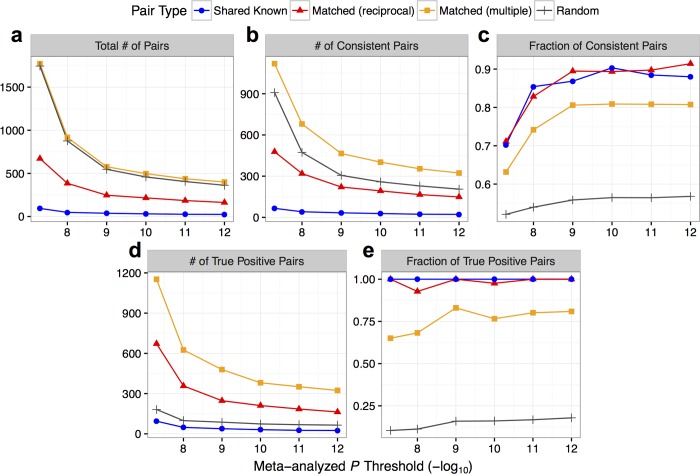
Fig 2. Genetic validation of OE-MCDS matched signals.
GWAS were performed for “multiple” matched signal pairs (a subset of which were “reciprocal”), shared known pairs (“Shared Known”, positive control), and randomly matched pairs (average statistics shown as “Random”, negative control) in OE and MCDS, followed by meta-analysis that ignored direction of effect. For each pair, the SNP with the best meta-analyzed p-value was selected to assess directional consistency of its association in the two cohorts. (a) “Total # of Pairs”: number of signal pairs with best SNPs below p-value threshold; (b) “# of Consistent Pairs” and (c) “Fraction of Consistent Pairs”: number and fraction of pairs with directionally consistent best SNPs below p-value threshold; (d) “# of True Positive Pairs” and (e) “Fraction of True Positive Pairs”: number and fraction of true positive pairs were estimated as described in Methods. X-axes start at genome-wide significant p-value threshold (p < 5 × 10−8). Error bars for “Random” pairs were excluded due to low visibility (all close to average).