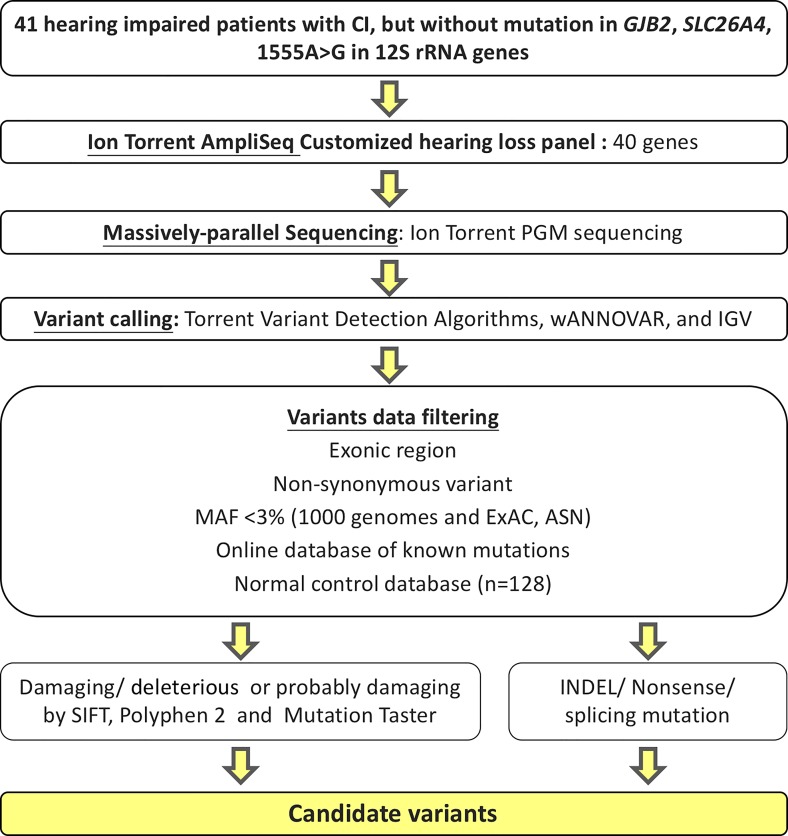
Fig 1. Pipeline used to identify candidate variants.
Forty-one patients with CI who lacked the known mutations in common deafness-associated genes were subjected to comprehensive genetic analysis by using an Ion Torrent PGM sequencer to target 40 relatively rare deafness genes. Variants were called using plug-in Torrent variant detection algorithms, annotated through wANNOVAR, and initially confirm using the integrative Genome viewer. Annotated variants were filtered by various criteria, including: being located in an exonic region; being non-synonymous; having an allele frequency < 3% in the 1000 Genomes Project; having a variant frequency of 30–70% for heterozygotes or >85% for homozygotes; and being absent from online databases and 128 ethnically matched normal hearing controls. SIFT, Polyphen 2, and Mutation taster were used to predict the functions of the identified variants; we first filtered for missense variants, and then directly identified indels, splicing site variants, and nonsense variants.