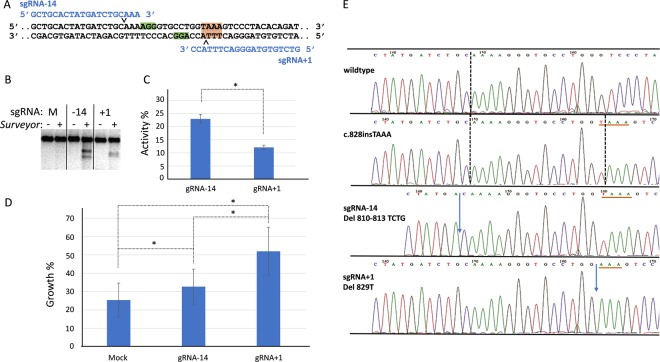
Figure 2.
CRISPR/Cas9 and error-prone repair activities restore Fancf function. (A) Design of sgRNAs to target Fancf c.828InsTAAA allele. Analysis of the Fancf c.828insTAAA DNA sequence revealed two opposing protospacer sequences (blue) with adjoining PAM motifs (green boxes) with predicted Cas9 nuclease activity sites at positions −14 (∨) and + 1(∧) of the c.828insTAAA mutation (Red box). (B) Surveyor assay indicates error-prone repair activity at targeted positions only when sgRNAs are present as measured by cleavage of heteroduplex DNA by the Surveyor nuclease. (C) Quantification of DNA cleavage by Surveyor (n = 3, one sided Student’s T-test, *p = 0.04). (D) Error-prone DNA end joining repair correlates with enhanced clonal survival in the presence of 15 nM MMC after double-strand break formation by Cas9 and sgRNA-14 or sgRNA + 1. (n = 3, one sided Student’s T-test, *p ≤ 0.04). (E) Fancf sequence analysis of single alleles from MMC-selected clones revealed rescued in-frame ORFs following DSB repair by mutagenic end joining. Del810–813 TCTG neutralizes the TAA stop codon at position 829, but also results in amino acid substitutions 271–276 LQKGAW > KRVPGK (see Suppl. Table 2A). While del829T restores the ORF, the FANCF peptide sequence contains an additional Lysine at position 279 (see Suppl. Table 2B). The location of the Fancf c.828insTAAA mutation is underlined in red. Vertical dotted lines indicate predicted Cas9 DSB sites. Blue arrows indicate the position of the deletions. Error bars represent standard error.