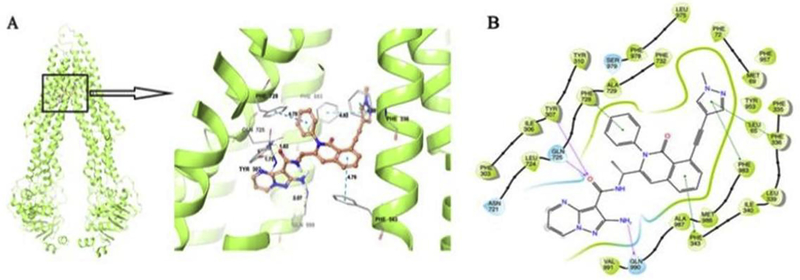
Figure 4. Induced Fit Docking (IFD) analysis of IPI-549 to ABCB1.
The best-scored binding pose of IPI-549 within human homology ABCB1 (Glide gscore: −14.602 kcal/mol) predicted by IFD computation is shown in (A). The location of the IPI-549 molecule as a ball and stick model is shown within the ABCB1 internal cavity, with the atoms colored as carbon–orange, hydrogen– white, oxygen–red, nitrogen–blue. Amino acids that have hydrogen bonding or π-π stacking interactions with IPI-549 are shown and depicted as sticks with the same color scheme as above except that carbon atoms are represented in grey. Only polar hydrogens are shown. Dotted yellow lines indicate hydrogen-bonding interactions, while dotted blue lines indicate π -π stacking interactions. Values of the relevant distances are given in Å. (B) The two-dimensional ligand−receptor interaction diagram of IPI-549 and human ABCB1. The amino acids within 4 Å are shown as colored bubbles, blue indicates polar residues, and green indicates hydrophobic residues. Grey circles indicate solvent exposure. Hydrogen bonds are shown by the purple arrow, and π-π stacking aromatic interactions are shown by green lines.