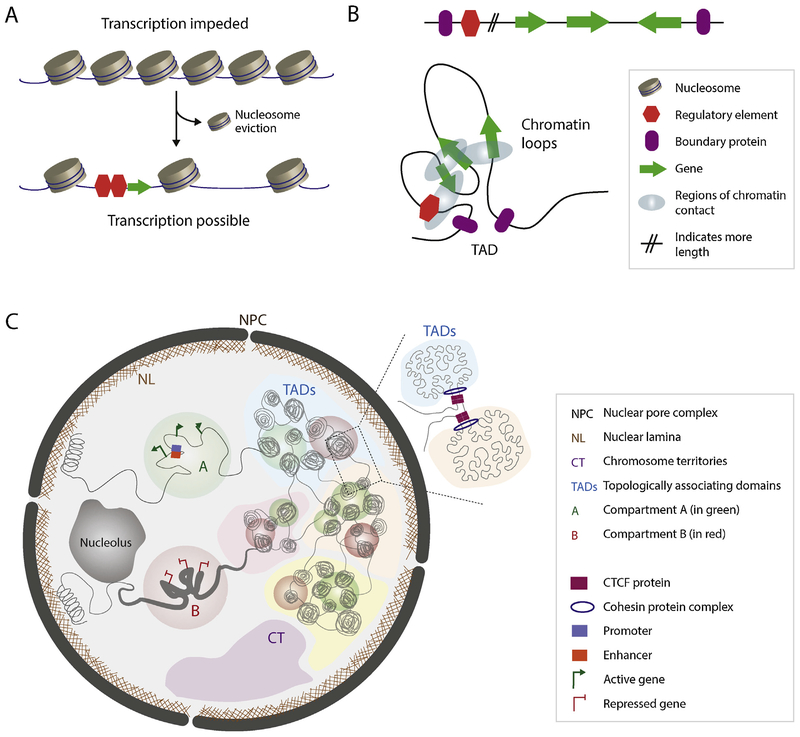
Fig. 1.
Hierarchical chromatin organization within the eukaryotic nucleus. (A) DNA is tightly wrapped around histone proteins and assembled into nucleosomes. This packaging system impedes transcription as transcriptional machinery cannot access the DNA. Local displacement of nucleosomes allows transcription machinery to access the genes. (B) Linear representation of a hypothetical genomic region. Possible chromatin loops that can form to specially connect promoters and their regulatory elements are depicted. (C) Model of 3D genome organization at different resolutions as described for animal models. During interphase chromosomes are organized into discrete chromosome territories (CTs). Within CTs, a particular locus can be surrounded by an active (A compartment) or repressive environment (B compartment). Different topologically-associating domains (TADs) with similar epigenetic signatures are characterized by strong inter-domain interactions and are organized into compartments. On the right, two different TADs are schematically represented. Insulator proteins (CTCF) are shown in purple rectangles and cohesin protein complexes are shown in blue circles.