Abstract
Inherent genetic programming and environmental factors affect fetal growth in utero. Epidemiologic data in growth-altered fetuses, either intrauterine growth restricted (IUGR) or large for gestational age (LGA), demonstrate that these newborns are at increased risk of cardiometabolic disease in adulthood. There is growing evidence that the in utero environment leads to epigenetic modification, contributing to eventual risk of developing heart disease or diabetes. In this study, we used reduced representation bisulfite sequencing to examine genome-wide DNA methylation variation in placental samples from offspring born IUGR, LGA, and appropriate for gestational age (AGA) and to identify differential methylation of genes important for conferring risk of cardiometabolic disease. We found that there were distinct methylation signatures for IUGR, LGA, and AGA groups and identified over 500 differentially methylated genes (DMGs) among these group comparisons. Functional and gene network analyses revealed expected relationships of DMGs to placental physiology and transport, but also identified novel pathways with biologic plausibility and potential clinical importance to cardiometabolic disease. Specific loci for DMGs of interest had methylation patterns that were strongly associated with anthropometric presentations. We further validated altered gene expression of these specific DMGs contributing to vascular and metabolic diseases (SLC36A1, PTPRN2, CASZ1, IL10), thereby establishing transcriptional effects toward assigning functional significance. Our results suggest that the gene expression and methylation state of the human placenta are related and sensitive to the intrauterine environment, as it affects fetal growth patterns. We speculate that these observed changes may affect risk for offspring in developing adult cardiometabolic disease.
Keywords: intrauterine growth restriction (IUGR), placenta, developmental programming of cardiometabolic disease, DNA methylation, large for gestational age (LGA)
Background
Fetal growth in utero is affected by inherent genetic potential in combination with environmental factors, such as maternal health and nutrition. Intrauterine growth restriction (IUGR) is a poorly understood complication of pregnancy, defined as growth less than genetically determined potential size, and affects 3% to 10% of pregnancies. The causes of IUGR are multiple but, in general, are thought to result from poor maternal nutrition (largely in poorly resourced countries) or poor nutrient provision to the fetus, which may result from disordered placentation (in well-resourced countries). Large for gestational age (LGA) newborns have birthweights greater than the 90th percentile for gestational age (GA). Large for gestational age is thought to result from genetic factors, maternal health conditions such as diabetes, or excessive maternal weight gain during pregnancy. In the short term, IUGR and LGA infants are both at increased risk of neonatal intensive care admission for management of hypo- or hyperglycemia, respiratory distress, or other conditions, necessitating prolonged postnatal monitoring.
In addition, epidemiologic data in growth-altered newborns, either IUGR or LGA, demonstrate compelling evidence that these offspring are at an increased risk of cardiovascular and metabolic disease in adulthood. It is widely accepted that environmental factors may be just as important as genetic predisposition in eventual risk of developing heart disease or diabetes.1 In fact, the influence of environment starts as early as conception. In utero exposure to altered nutrient provision, as affected by maternal diet or pregnancy complications, sets the stage for how the human body can respond to environmental stress and food excess postnatally, and subsequently in adulthood. This phenomenon, by which the environment of pregnancy confers risk of future adult disease to the developing fetus, is known as “developmental programming.”
Although we now recognize the importance of early environment on risk for disease in later life,2–6 we still have a poor understanding of the mechanisms that underlie this phenomenon. The study of epigenetics provides mechanisms that allow for interaction between genetic and environmental influences that lead to risk for adult disease. The placenta is a uniquely suited organ for the study of how genetics (fetal cells) and environment (eg, maternal health and nutrient provision) interact and may provide considerable insights into the developmental programming of disease. Others and we have described in both humans and in animal models that there is evidence of altered gene expression and epigenetic regulation of key players in cardiovascular and metabolic pathways in growth-altered fetuses.7–9 As most of these reports are based on candidate gene analysis or low-resolution array data, there is still a paucity of genome-wide methylation analysis relevant to this specific topic of fetal growth patterning. Although the existing studies are important and provide insight into specific processes, a genome-wide approach allows for both perspective into larger-scale complex processes of genomic regulation without the bias of specific pathways of interest and the ability to identify individual cardiometabolic genes in which differential methylation may confer risk for later disease.
Our study is a discovery-based investigation of genome-wide epigenetic variation in fetal tissues based on prenatal growth patterns and birthweight. We hypothesized that altered fetal growth, either IUGR or LGA fetuses, may correlate with changes in placental gene expression and DNA methylation in genes conferring risk for cardiometabolic disease. Both the fetus and placenta are simultaneously exposed to the same maternal intrauterine environment. Thus, unique placental epigenomic signatures based upon in utero growth patterns, with changes that have functional importance to human disease, may serve as biomarkers for adult cardiometabolic phenotypes. To test this hypothesis, we isolated DNA and RNA from human placentas taken from term pregnancies resulting in appropriate for gestational age (AGA) growth, IUGR, and LGA infants based upon intrauterine growth patterns. We profiled DNA methylation genome-wide and performed pathway analysis to identify differentially methylated regions located in selected transcriptionally important regions of key and significant genes in cardiovascular and metabolic pathways. We independently validated our data using a data set of differentially methylated regions (DMRs) in human placentas in a larger cohort of participants. In order to establish biologic importance and transcriptional effect to methylation changes, by real-time quantitative reverse transcription polymerase chain reaction (qRT-PCR), we further validated alterations in the expression of selected genes that mediate cardiometabolic risk and were differentially methylated among groups.
Materials and Methods
Human Placental Samples
Collection and processing of the placentas used for this study have been described previously.10 Briefly, informed consent was obtained from women with AGA fetuses (n = 6), with LGA fetuses (n = 5), or with late-onset IUGR (n = 6). Large for gestational age was defined by estimated fetal weight ≥∼90th percentile for GA, and IUGR was defined by estimated fetal weight ≤10th percentile for GA and a trajectory of fetal growth deceleration in utero, diagnosed by ultrasound and intrauterine growth curves. Given the myriad causes of IUGR, we specifically selected cases of “idiopathic” IUGR in order to minimize clinical heterogeneity in this group. Mothers with gestational or chronic hypertension, obesity, or reported drug use (specifically smoking) were excluded. Similarly, for LGA cases, mothers with gestational or pregestational diabetes were excluded. Of note, 1 mother in the LGA cohort had a diagnosis of obesity. The decidual layer, basal plate, and chorionic surface and membranes were removed by sharp dissection, and placental fragments were obtained at the middle of the initial placental depth. Placental samples were snap frozen and stored at −80°C.
Reduced Representation Bisulfite Sequencing
Genomic DNA from human placentas was extracted for constructing reduced representation bisulfite sequencing (RRBS) libraries following the standard RRBS protocol.11 Genomic DNA was digested with a methylation-insensitive endonuclease, MspI. Fragments from 40 to 220 base pairs (bp) were isolated as they are enriched for CpG-rich regions, such as CpG islands and promoter regions. The MspI-digested DNA was end-repaired, A-tailed, and ligated with Illumina adaptors. The double-stranded DNA (dsDNA) was denatured followed by bisulfite conversion and PCR amplification. These libraries were sequenced with Illumina Hiseq 2000 sequencers. The reads were aligned to the reference genome (human hg19) using the bisulfite aligner, BS Seeker2,12 and only uniquely mapped reads were kept. The overall bisulfite conversion rate was 97.3% (Supplementary File 1). With BS Seeker 2, we calculated the methylation level for each cytosine (Cs) on the Mspl-digested restricted genome. As bisulfite treatment converted unmethylated Cs to thymines, the methylation level at each Cs was estimated by #C/(#C+#T), where #C is the number of methylated reads and #T is the number of unmethylated reads. In this study, only Cs that are covered by at least 4 reads were included for subsequent analysis, which includes 75% of the Cs covered by our reads. We did not consider single-nucleotide polymorphisms (SNPs) in our analysis, but we have examined other human RRBS data and found that only 300 to 600 C/T SNPs are found within the RRBS fragments. Compared to our coverage of more than 4M CpG sites, SNPs likely affect only 0.0015% of the Cs.
Identifying Differentially Methylated Regions and the Associated Genes
We first searched for DMR that showed significant differential methylation. Genes that were close to these DMR were considered differentially methylated. For each cytosine-guanine base pairing (CG site), we calculated a t score from the t test of mean difference between the 2 groups of comparison, then selected sites with |t score| ≥ 1.5 (accounting for approximately the top 10%) as markers of differential methylation. If 2 markers were within 80 bp (in our data, median distance = 74 bp), then the region between them was deemed a candidate DMR. For each candidate DMR, we then calculated a z score of the average t score from all CG sites within the region, as a measure of the differential methylation within the candidate DMR. When the |z score| was greater than a threshold and the mean methylation levels in the 2 groups differed by at least 15%, this region was considered as a DMR. Each DMR had to contain at least 4 common CpG sites that have methylation data available for the samples between the 2 compared groups. Finally, if genes overlapped with any of these DMRs or if their transcription start sites were within 5 kb of the DMR, these genes were deemed differentially methylated. Approximately 55% of the DMR were proximal to genes, and 26% of the DMRs were associated with more than 1 gene.
Estimating False Discovery Rate in DMR Identification
To assess the false discovery rate (FDR), we constructed 17 simulated methylomes, with the same sequencing depth per site as the real samples. For each CG site, in each simulated sample, we then simulated the bisulfite converted reads (C if methylated or T if unmethylated) based on the average methylation level (Pm), which was estimated from all real samples at this CG site. We assume that the number of methylated reads (Cs) at a site of depth n is a random variable following a binomial distribution B(n,Pm). We repeated our simulation of reads throughout the genome for all samples. The resulting methylomes would have the same average methylation levels per CG site as the real sample. Since the reads were simulated from the binomial distribution with the same average methylation levels (Pm) as in the real samples, the differences in methylation patterns across genes, repeats, promoters, and so on, were preserved. The simulated data also had the same coverage as the real samples so the statistical power for detecting DMR is the same between the real and simulated samples. Moreover, the simulated methylomes should have no difference in methylation levels between any 2 groups that are compared (ie, no DMR), since they are all constructed from the same parameter Pm. Therefore, any DMR (and the DMR associated genes) identified from these simulated methylomes are thus considered false positives, and the DMR detected from the real sample include both true positives and false positives. Finally, the FDR is estimated as the ratio between the number of false positives and the number of all positives (true positives and false positives). We selected DMR with the criteria |Δm| > 15% such that FDR was ≤5%.
Covariance and t Test
For each CG site in the 17 RRBS libraries with coverage >4X, we calculated the covariance between methylation level and individual phenotypes, that is, birthweight, length, head circumference (HC), and GA. To minimize false positives, we selected data with a FDR ≤ 10%, and the criteria were as follows: GA, covariance >1.008 and <−0.964; HC, covariance >0.232 and <−0.208; length, covariance >0.434 and <−0.386; weight, covariance >100.8 and <−93.0. To evaluate the relationship between mode of delivery and methylation level, for each CG site in the 17 RRBS libraries with coverage >4X, we performed a t test between cesarean section (C/S) and normal spontaneous vaginal delivery (NSVD), selected sites that had |t score | > 10, and calculated the differential methylation level (Δm) per site (C/S vs NSVD). To minimize false positives, we generated 17 simulated CG maps, which have similar methylation level on each site, and FDR is based on comparing Δm between real and simulated methylomes. We selected data with the criteria Δm > 0.22 and Δm < −0.24 such that FDR ≤ 10% and performed Genomic Regions Enrichment of Annotations Tool (GREAT) functional analysis. We included in Supplementary File 2 both lists of genes with high covariance sites from FDR ≤5% and FDR from ≤10%.
Analysis of Variance
For each CG site in the 17 RRBS libraries that has coverage more than 4, we calculated the F value from the analysis of variance (ANOVA) of mean difference among AGA, LGA, and IUGR groups of comparison and sorted the F values in descending order. We selected 1000 CG sites with the highest F values and submitted them to the GREAT analysis tools to predict functional pathways.13
Functional Pathway and Network Analysis of DMRs and of Differentially Methylated Genes
To understand the functional pathways and networks affected by our DMR and differentially methylated genes (DMG), we performed GREAT13 and STRING14 analyses. Genomic Regions Enrichment of Annotations Tool (GREAT) explores functional categories likely to associate with DMR by analyzing the annotations of nearby genes. The analysis by STRING presents the network structure of DMG to identify the potential interacting partners and the enriched functional modules. Both of these programs utilize curated data sets that are generated from both published information and computational predictions.
Gene and Protein Expression of DMGs in the Placenta Using GeneCards
In order to establish the biologic plausibility of our DMGs to change the placental transcriptome, we examined previously reported messenger RNA (mRNA) expression of our DMG using the GeneCards database.15 We sorted the top DMG for each comparison (by Z score) and evaluated reported placental expression for the DMGs associated with the top 50 DMRs.
Real-Time Quantitative PCR
For RT-PCR analysis, total cellular RNAs were isolated using the RNeasy mini kit (Qiagen, Valencia, California), according to the manufacturers’ instructions. Quantitative and qualitative analyses of isolated RNA were assessed by the ratio of absorbance at 260 and 280 nm. Complementary DNA was generated from 1 μg of total RNA from placental tissue using reverse transcription (RT) using a Superscript III Reverse Transcriptase kit (Invitrogen, San Diego, California), following the manufacturer’s instructions. The RT was performed at 50°C using 5μg/μL of random hexamers. Real-time PCR amplification was performed in triplicate using Taqman-based detection according to the manufacturer’s instructions on a Step One real-time quantitative PCR thermocycler (Applied Biosystems, Foster City, California). For each gene of interest, a Fam/Tamra probe was used (Eurofins, Louisville, KY). Relative gene expression was calculated using the comparative CT method16 with 18S (Applied Biosystems, #4319413E) expression used as the internal control for normalization. The amplification cycles consisted of 50°C for 2 minutes, 95°C for 20 seconds, then 45 cycles of 95°C for 1 second (denaturation), annealing for 20 seconds. Annealing temperatures, exon-spanning primers for amplification, and probe sequences for detection are included in Supplementary File 3.
Data Access
The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE70364.
Results
To investigate the effects of birthweight and fetal growth patterns on placental methylation of cardiometabolic genes, we isolated DNA from 17 whole placental samples (5 from pregnancies predicting an LGA infant, 6 from pregnancies complicated by IUGR, and 6 from pregnancies predicting an AGA infant based on prenatal ultrasound information). We also collected clinical and phenotypic data for each sample, including gestational age and gender of the newborn, delivery method, as well as weight, length, and HC of the newborn (Table 1). This study received institutional review board approval from the University of California–Los Angeles, and placentas and clinical information were collected only after obtaining maternal consents.
Table 1.
Clinical Phenotypes of All Samples Used for RRBS.a
| Weight | Length | HC | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Samples | GAb | Sexc | Deliveryd | g | Percentile Among Newborns | cm | Percentile Among Newborns | cm | Percentile Among Newborns | PIe |
| AGA | ||||||||||
| C1 | 41 + 5 | M | NSVD | 3795 | 40% | 55.9 | 95% | 34.3 | 10% | 2.173 |
| C4 | 39 + 5 | M | NSVD | 3260 | 30% | 50.8 | 50% | 32.0 | 5% | 2.487 |
| C7 | 39 + 4 | F | C/S | 3940 | 80% | 52.1 | 75% | 35.6 | 50% | 2.786 |
| C14 | 39 | F | C/S | 3460 | 50% | 51.0 | 60% | 35.0 | 50% | 2.608 |
| C16 | 39 + 6 | M | C/S | 4054 | 75% | 52.1 | 75% | 34.3 | 25% | 2.867 |
| C25 | 39 + 2 | F | C/S | 3275 | 40% | 48.3 | 25% | 35.6 | 50% | 2.906 |
| LGA | ||||||||||
| L1 | 41 + 5 | M | C/S | 4655 | >90% | 54.6 | 80% | 36.0 | 40% | 2.860 |
| L2 | 41 + 2 | M | C/S | 4430 | 85% | 53.0 | 75% | 36.0 | 40% | 2.976 |
| L4 | 40 + 6 | F | NSVD | 4170 | 86% | 53.34 | 84% | 33.02 | 6% | 2.748 |
| L6 | 40 + 0 | F | NSVD | 4256 | 94% | 58.42 | 100% | 33.655 | 22% | 2.135 |
| L7 | 40 + 3 | M | NSVD | 4564 | 96% | 55.88 | 18% | 33.999 | 97% | 2.616 |
| IUGR | ||||||||||
| U2 | 37 + 6 | F | NSVD | 2475 | 8% | 48.3 | 40% | 33.0 | 10% | 2.197 |
| U3 | 37 + 5 | M | NSVD | 2268 | 3% | 46.0 | 40% | 31.0 | 5% | 2.330 |
| U4 | 38 + 4 | M | C/S | 2551.5 | 5% | 49.5 | 50% | 33.0 | 20% | 2.104 |
| U5 | 38 | F | C/S | 2190 | <3% | 47.0 | 15% | 30.5 | 3% | 2.109 |
| U8 | 40 + 2 | M | C/S | 2580 | <3% | 46.5 | <3% | 30.9 | <3% | 2.566 |
| U10 | 39 + 5 | F | NSVD | 2863 | 9% | 48.3 | 15% | 30.5 | <3% | 2.541 |
| ANOVA between phenotypes and grouping F-value (P value) | 7.432 (0.006) | 69.314 (<0.001) | 18.211 (<0.001) | 10.400 (<0.005) | 3.386 (0.063) | |||||
Abbreviation: AGA, appropriate for gestational age; GA, gestational age; HC, head circumference; NSVD, normal spontaneous vaginal delivery.
aTable includes clinical information such as gestational age (GA), sex of infant, mode of delivery, weight, length, head circumference, and Ponderal index.
bGA= gestational age (weeks + days).
cSex: M = male; F = female.
dDelivery: NSVD = normal spontaneous vaginal delivery; C/S:= cesarean section.
ePI = Ponderal index = weight/height3 × 100.
These samples were subjected to genome-wide DNA methylation profiling by RRBS. This approach involves a restriction digest followed by size selection to enrich a genomic fraction for CpG-rich regions. On average, for each sample, we sequenced 46M reads with a mapability of ∼56% and sequencing depth of 13.3X (Supplementary File 1). This allowed us to measure methylation levels (from 0 = unmethylated, to 100% = fully methylated) of ∼4.3M CpG sites enriched in CpG islands.
We first compared the DNA methylation patterns among LGA, IUGR, and AGA placentas, to determine whether there were both global and gene level changes of methylation that were associated with fetal growth pattern and birthweight (AGA vs IUGR vs LGA).2 We then examined the correlation between DNA methylation and the phenotype data collected on each sample by performing covariance analysis using all individual Cs, to both (1) ensure that other clinical factors other than anthropometric measurements did not significantly affect placental methylation, and (2) to identify specific cardiometabolic genes that showed strong covariance with growth parameters.
Global Methylation Patterns Differ Among IUGR, LGA, and AGA Groups
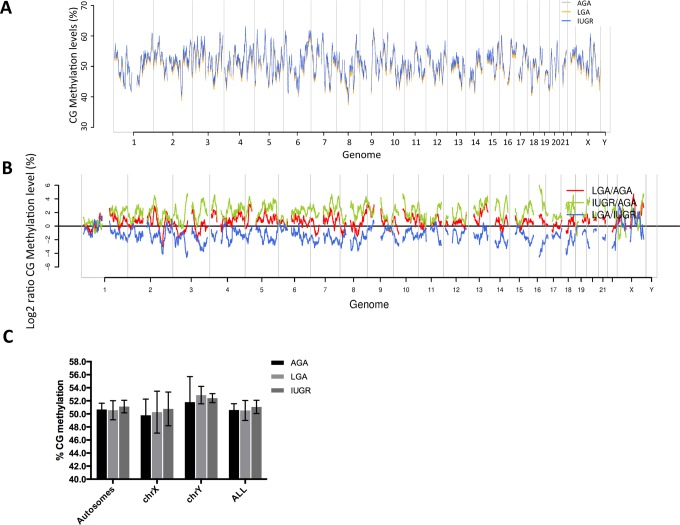
We began by comparing the average methylation level of placentas from different birthweight groups. Overall, the average CpG methylation level is around 50% in all groups with no significant differences between groups (Figure 1; Supplementary File 4). In addition, we found that placental methylation levels are not different between males and females, except for chromosome X (Supplementary File 5), and there is no significant difference between the groups within the same sex (Supplementary File 6 and 7). The three birthweight groups show similar methylation patterns in coding genes and transposons as in the genome-wide view (Supplementary File 8). However, this pattern changes in CpG islands where placentas associated with IUGR become less methylated than LGA and AGA placentas (Supplementary File 8B). This observation is in line with our previous murine observations7 and suggests a redistribution of methylation away from CpG islands to genes and transposons in IUGR.
Figure 1.
Profiling of genome wide DNA methylation in human placenta. A, Genome-wide cytosine-guanine pairing methylation levels (%) in AGA (gray), LGA (yellow), and IUGR (blue) groups. B, Log2 ratios of CG methylation level (%) in pair-wise comparisons (red: LGA vs AGA, green: IUGR vs AGA, blue: LGA vs IUGR). C, Average CG methylation levels in autosomes and sex chromosomes (dark gray: AGA, light gray: LGA, medium gray: IUGR). AGA indicates appropriate for gestational age; IUGR, intrauterine growth restriction; LGA, large for gestational age.
To identify the genomic regions that are susceptible to changes in intrauterine environment resulting in fetal growth alteration, we performed a genome-wide screen for DMRs (Δ methylation level ≥15% and FDR <5%; see Methods for DMR identification and FDR estimation). We identified 1015, 906, and 1022 DMRs, corresponding to 583, 516, and 567 genes that showed differential methylation in the comparisons between IUGR versus AGA, LGA versus AGA, and LGA versus IUGR (see Supplementary File 9 for a list of DMR and the associated genes). Using these criteria, we selected approximately 1% of the regions and 0.5% of the genes covered by RRBS data as differentially methylated.
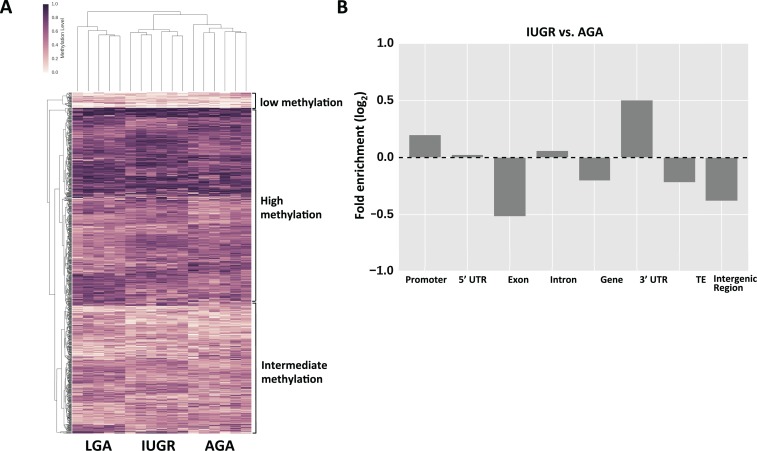
We found that the methylation patterns of DMR show unique signatures of methylation for each birthweight group, such that these patterns distinguish the appropriate intrauterine exposure group (based on the associated neonatal birthweight category) of each of our placental samples (Figure 2A). This result suggests that in these regions there is an association between the methylation pattern and intrauterine environment affecting birthweight. In Supplementary File 10, we provide a list of the top 1000 Cs whose methylation levels are associated with infant size groups based on ANOVA.
Figure 2.
Differentially methylated regions. A, Heatmap of methylation variable regions across LGA, IUGR, and AGA groups. Darker color denotes higher methylation levels, while lighter colors denote low methylation levels. B, Enrichment analysis of DMR location between IUGR and AGA. Similar patterns were seen comparing LGA and AGA groups (data not shown). AGA indicates appropriate for gestational age; DMR, differentially methylated region; IUGR, intrauterine growth restriction; LGA, large for gestational age.
We also noticed from the dendrogram (Figure 2A) that these DMRs are clustered as groups of high, medium, and low average methylation (with 513, 330, and 39 DMRs, respectively). The functional analysis of the high and low methylation DMRs shows that they are enriched in different functional categories (Supplementary File 11).
In general, the DMRs were found to be more abundant in promoters and three prime untranslated region (3′UTR) and depleted from coding regions, such as exons, gene bodies, transposable elements, as well as intergenic regions (Figure 2B; Supplementary File 12). The genes associated with DMR were considered differently methylated genes and were subjected to gene network analysis using the Ingenuity Pathway Analysis tool (Ingenuity Pathway Analysis software, Ingenuity Systems, California). These genes were found to be enriched in specific networks responsible for both cardiovascular disease and metabolic disorders (when comparing IUGR or LGA vs AGA). Other networks identified included cell-to-cell signaling and interaction, connective tissue development and function, and cellular assembly and organization, as expected (see Table 2 for top networks).
Table 2.
Differentially Methylated Genes and the Enriched Gene Networks.a
| Comparison | #DMR | #Differentially Methylated Genes | Top Major Networks (Significance Score = −log10[P value]) |
|---|---|---|---|
| IUGR vs AGA | 1015 | 191 |
|
| LGA vs AGA | 906 | 171 |
|
| LGA vs IUGR | 1022 | 172 |
|
Abbreviations: AGA, appropriate for gestational age; DMR, differentially methylated region; IL10, interleukin 10; IUGR, intrauterine growth restriction; LGA, large for gestational age.
aThe number of DMR and DMR genes found between each comparison group and the top major networks represented by those DMR.
Independent Validation of DMR
As an independent validation of our DMR analysis, we reanalyzed the published methylation array data from Marsit et al,8 in which DNA methylation of human placentas was compared between IUGR/SGA and AGA using the Illumina Infinium HumanMethylation27 BeadChip array in a large cohort of 206 human placentas. The 27K array has much less coverage compared to RRBS, which covers 4.3M CpG sites. The data we retrieved from their 27K array are a matrix of log2 ratios between methylated and unmethylated probes. We first looked for probes on the array that are within our DMRs measured across the 3 comparison groups (IUGR-AGA, LGA-IUGR, and LGA-AGA). As our DMRs are small, with an average size of 73 to 75 bp, there are only 3 that are covered by a probe from the 27K array data. In these 3 cases, we tested whether the probe within our DMR shows differential methylation between the comparison groups in the cohort of Marsit.8 We used both parametric (t test) and nonparametric (Mann-Whitney U test and Kolmogorov-Smirnov test) tests (Table 3) to determine the significance of the differences. Using this approach, we are able to validate 2 of the 3 DMRs (2 DMRs from IUGR-AGA and 1 DMR from LGA-IUGR). All 3 probes showed the same sign of methylation differences (ie, the differences between groups were in the same direction), and 2 reached statistical significance.
Table 3.
Validation of Differentially Methylated Regions Identified by RRBS With 450K Methylation Array Data.8,a
| Comparison DMR (RRBS) | Δ CpG methylation | Probe_ID | Distance to DMR | t-Test Score | t-Test: P Value | Hyper/Hypostatus | Mann-Whitney: P Value | Kolmogorov-Smirnov Test: P Value |
|---|---|---|---|---|---|---|---|---|
| IUGR-AGA | ||||||||
| chr5:175969381-175969447 | −0.207 | cg22733478 | 0 | −2.080 | 0.040b | Consistent | .014b | .066 |
| chr3:185912389-185912478 | −0.255 | cg24490859 | 0 | −2.623 | 0.010b | Consistent | .005b | .011b |
| LGA-IUGR | ||||||||
| chr7:989199-989229 | 0.248 | cg15914863 | 0 | 1.381 | 0.198 | Consistent | .071 | .058 |
Abbreviations: AGA, appropriate for gestational age; DMR, differentially methylated region; IUGR, intrauterine growth restriction; LGA, large for gestational age; RRBS, reduced representation bisulfite sequencing.
aTable including 3 loci where methylation levels were measured by both methods, the difference in methylation levels between groups, t-test score, and p values.
bSignificance with p-value < 0.05 by t-test.
Specific Loci Show Strong Covariance With Clinical Phenotypes in Offspring
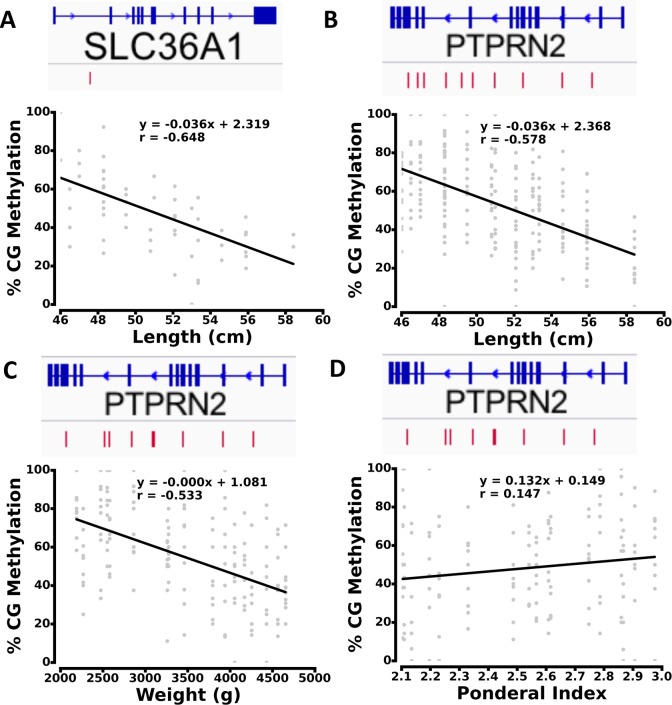
To identify specific loci whose methylation level had a strong association with the individual fetal growth and newborn phenotypes, we utilized additional individual clinical anthropometric measurements (length, HC) beyond birthweight alone. We also examined other important clinical characteristics (such as mode of delivery and gestational age) to distinguish effects that may be related to those factors. For each C, we calculated the covariance between its methylation levels and the phenotype across 17 placental samples. Depending on the phenotypes, approximately 2000 CpG sites (FDR < 10%) were found to be highly correlated with the clinical phenotypes (see Methods for covariance analysis and FDR estimate). To visualize high covariance sites across genes, we generated scatter plots of the methylation levels at these Cs versus the phenotype values (see Figure 3). For example, the methylation levels of CpGs associated with SLC36A1 showed clear correlation with the length of the offspring. (Figure 3A). Within PTPRN2, the methylation at a specific set of Cs had an inverse relationship with both length (Figure 3B) and weight (Figure 3C) of the offspring. We also calculated Ponderal index (PI = 100 × (weight/length3)), using weight in grams and length in cm, and demonstrated that for PTPRN2, PI shows a linear relationship with CG methylation levels at high covariance sites (Figure 3D).
Figure 3.
Scatter plot of high covariance methylation sites with phenotypes. Covariance between methylation sites and length in: (A) SLC36A1, and (B) PTPRN2. C, Covariance between methylation sites and weight in PTPRN2. D, Covariance between methylation sites and Ponderal index in PTPRN2. The red bars in the gene models show the distribution of high covariance cytosines.
We performed a functional analysis of genes with high covariance Cs associated with different phenotypes (Table 4). The enriched categories include protein transport, glucose and glycogen metabolism, and hormone synthesis, processes which are key to maintaining nutrient provision and growth signaling to the fetus, especially when considering growth phenotypes (weight, length, HC, and PI). Mode of delivery and gestational age demonstrated few to no enriched processes associated with cardiometabolic disease.
Table 4.
Gene Otology of Phenotype-Associating Genes From Covariance Analysis.a
| Phenotype | Function Group | Fold Enrichment | Corrected P Value | GO Biological Process |
|---|---|---|---|---|
| GA | Molecular function | 16.7150 | 6.40 × e−12 | Calcium-dependent phospholipase C activity |
| 7.1453 | 1.50 × e−7 | Protein-tyrosine sulfotransferase activity | ||
| Biological process | 7.1453 | 1.50 × e−7 | Peptidyl–tyrosine sulfation | |
| 2.9369 | 2.67 × e−6 | Neutral amino acid transport | ||
| Cellular component | 10.7063 | 4.00 × e−6 | Alpha6-beta4 integrin complex | |
| 13.5658 | 1.96 × e−4 | Nuclear RNA-directed RNA polymerase complex | ||
| Human phenotype | 16.0007 | 3.12 × e−8 | Joint contractures, progressive | |
| 4.6027 | 5.76 × e−8 | Abnormality of the tendons | ||
| Weight | Molecular function | 22.3820 | 1.24 × e−13 | Phenylpyruvate tautomerase activity |
| 10.1030 | 3.03 × e−11 | Glycerol kinase activity | ||
| Biological process | 3.1119 | 6.45 × e−10 | Protein trimerization | |
| 3.5484 | 1.43 × e−9 | Protein homodimerization | ||
| Human phenotype | 21.3946 | 2.30 × e−16 | Joint contractures, progressive | |
| 10.2936 | 2.30 × e−11 | Thickened skin | ||
| Length | Molecular function | 4.8631 | 8.16 × e−9 | Low voltage-gated calcium activity |
| 5.5497 | 3.86 × e−8 | Protein-tyrosine sulfotransferase activity | ||
| Biological process | 7.0935 | 9.00 × e−12 | Aldosterone biosynthetic process | |
| 7.0935 | 9.00 × e−12 | Cortisol biosynthetic process | ||
| Human phenotype | 3.1737 | 2.70 × e−6 | Abnormality of tendons | |
| HC | Molecular function | 11.1799 | 1.84 × e−15 | Glycerol kinase activity |
| 14.1612 | 4.09 × e−11 | Calcium-dependent phospholipase C activity | ||
| Biological process | 25.0350 | 2.35 × e−13 | Leptotene | |
| 7.4178 | 4.68 × e−13 | Glycerol-3-phosphate metabolic process | ||
| Human phenotype | 21.6211 | 1.22 × e−20 | Joint contractures, progressive | |
| 11.7389 | 1.30 × e−16 | Thickened skin | ||
| Delivery | Molecular function | 16.5429 | 1.14 × e−5 | Filamin-A binding |
| Biological process | 7.9001 | 2.38 × e−6 | Alveolar secondary septum development | |
| Human phenotype | 8.3005 | 1.11 × e−7 | Capitate-hamate fusion | |
| 5.2453 | 2.72 × e−7 | Periorbital fullness | ||
| PI | Molecular function | 26.5879 | 6.72 × e−47 | Glycerol kinase activity |
| 16.3202 | 2.24 × e−7 | Formimidoyltetrahydrofolate cyclodeaminase activity | ||
| Biological process | 18.0846 | 3.06 × e−39 | Glycerol-3-phosphate metabolic process | |
| 7.2580 | 1.76 × e−25 | Glycerol metabolic process | ||
| Cellular component | 2.1967 | 6.20 × e−9 | Mitochondrial outer membrane | |
| 6.2218 | 2.05 × e−6 | Integral to nuclear inner membrane | ||
| Human phenotype | 42.7104 | 2.70 × e−57 | Joint contractures, progressive | |
| 27.4899 | 8.75 × e−50 | Thickened skin |
Abbreviations: HC, head circumference; GA, gestational age; PI, Ponderal index; GO, gene ontology.
aGenes demonstrating high covariance by methylation level with clinical phenotypes.
Functional Pathway and Gene Network Analysis of DMRs and of DMGs
To understand the functional pathways and networks associated with our DMRs in comparisons between AGA, IUGR, and LGA, we used GREAT analysis. We found that our sets of DMRs predict pathways involving placental physiology and transport (Table 5) as expected, which lends biologic plausibility to the findings in our study. In addition, there are a number of interesting pathways highlighted by GREAT analysis that may be important for growth and cardiometabolic pathways, such as the protein RNF187 pathway. This protein encodes a ligase that acts as a coactivator of JUN-mediated gene activation in response to growth factor signaling via MAP3K1. However, GREAT analysis also uncovers novel pathways such as (1) cancer-related pathways, especially in the LGA comparison, which also represent a category of overgrowth syndromes, (2) endocrine pathways such as thyroid stimulating hormone, insulin receptor signaling, and glycogen synthase pathways, and (3) neurologic pathways (eg, myelin). These novel findings can be used for future hypothesis generation and subsequent studies.
Table 5.
Genomic Regions Enrichment of Annotations Tool (GREAT) Analyses.a
| Comparison | Hyper Raw P Value | Hyper FDR Q-val | Hyper Fold Enrichment | Significant Functional Pathways |
|---|---|---|---|---|
| IUGR vs. AGA | 4.9137 × e− 5 | 5.5279 × e− 2 | 4.9320 | Podosome (GO cellular component) |
| 3.0438 × e− 7 | 2.4299 × e− 3 | 16.4984 | Protein RNF187 (TreeFam) | |
| 3.3197 × e− 6 | 1.3250 × e− 2 | 11.5156 | Rho-related GTP-binding protein RhoV RhoU (Tree Fam) | |
| LGA vs. AGA | 2.6922 × e− 5 | 3.0287 × e− 2 | 2.6931 | Neuron projection terminus (GO cellular component) |
| 6.9863 × e− 5 | 3.9298 × e− 2 | 2.6610 | Axon terminus (GO cellular component) | |
| 3.0930 × e− 6 | 2.2557 × e− 2 | 5.4859 | Abnormal placental physiology (mouse phenotype) | |
| 8.8175 × e− 6 | 3.2153 × e− 2 | 4.5579 | Abnormal extraembryonic tissue physiology (mouse phenotype) | |
| 2.1239 × e− 5 | 5.1632 × e− 2 | 56.7979 | Abnormal organ of Corti supporting cell proliferation (mouse phenotype) | |
| 2.6665 × e− 5 | 4.8617 × e− 2 | 8.3369 | Abnormal placental transport (mouse phenotype) | |
| 3.1784 × e− 5 | 4.6360 × e− 2 | 8.1083 | Enlarged inguinal lymph nodes (mouse phenotype) | |
| 3.7734 × e− 5 | 4.5866 × e− 2 | 47.0611 | Increased organ of Corti supporting cell number (mouse phenotype) | |
| 3.7981 × e− 5 | 3.9571 × e− 2 | 4.2479 | Increased thyroid-stimulating hormone level (mouse phenotype) | |
| 4.2729 × e− 5 | 3.8953 × e− 2 | 4.5547 | Increased circulating thyroid-stimulating hormone level (mouse phenotype) | |
| 4.5548 × e− 5 | 3.6909 × e− 2 | 3.8834 | Maternal imprinting (Mouse phenotype) | |
| 2.1239 × e− 5 | 4.7427 × e− 2 | 56.7979 | Familial retinoblastoma, bilateral retinoblastoma (disease ontology) | |
| LGA vs. IUGR | 2.7542 × e− 7 | 9.3671 × e− 4 | 10.5601 | CTD phosphatase activity (GO molecular function) |
| 1.8216 × e− 6 | 1.5959 × e− 2 | 16.9861 | Insulin receptor signaling pathway via phosphatidylinositol 3-kinase cascade (GO biological process) | |
| 5.8649 × e− 6 | 2.5691 × e− 2 | 10.5528 | Gonadal mesoderm development (GO biological process) | |
| 1.0997 × e− 5 | 3.2115 × e− 2 | 9.5746 | Regulation of glycogen (starch) synthase activity (GO biological process) | |
| 2.2774 × e− 5 | 4.9881 × e− 2 | 10.9015 | Positive regulation of glycogen (starch) synthase activity (GO biological process) | |
| 2.5493 × e− 5 | 4.4669 × e− 2 | 6.0198 | Fibril organization (GO biological process) | |
| 5.7476 × e− 6 | 2.0958 × e− 2 | 85.9298 | Decreased activity of parathyroid (mouse phenotype) | |
| 6.2276 × e− 6 | 1.5139 × e− 2 | 10.4557 | Absent pulmonary valve, aortic valve cusps (mouse phenotype) | |
| 6.6773 × e− 6 | 9.7396 × e− 3 | 8.4136 | Abnormal placental transport (mouse phenotype) | |
| 2.2514 × e−5 | 2.7366 × e−2 | 5.4164 | Hypocalcemia (mouse phenotype) | |
| 2.5252 × e− 5 | 2.6309 × e− 2 | 10.7019 | Delayed eyelid fusion (mouse phenotype) | |
| 3.0255 × e− 5 | 2.7582 × e− 2 | 10.3603 | Abnormal compact bone lamellar structure (mouse phenotype) | |
| 4.2905 × e− 5 | 3.4767 × e− 2 | 2.4935 | Abnormal lens development (mouse phenotype) | |
| 6.7228 × e− 5 | 4.9029 × e− 2 | 7.1910 | Abnormal nucleus accumbens morphology (mouse phenotype) | |
| 4.7763 × e− 8 | 2.8572 × e− 4 | 13.0042 | Decreased serum estradiol (human phenotype) | |
| 4.7763 × e− 8 | 2.8572 × e− 4 | 13.0042 | Rhabdomyolysis, acute (human phenotype) | |
| 5.5481 × e− 8 | 1.1063 × e− 4 | 12.7767 | Prominent midface (human phenotype) | |
| 5.1024 × e− 7 | 7.6306 × e− 4 | 9.8041 | Peripheral hypomyelination (human phenotype) | |
| 1.1858 × e− 6 | 1.4186 × e− 3 | 4.1977 | Microcornia (human phenotype) | |
| 1.6302 × e− 6 | 1.6253 × e− 3 | 8.5096 | Genu recurvatum (human phenotype) | |
| 4.7590 × e− 6 | 4.0669 × e− 3 | 3.7764 | Congenital cataract (human phenotype) | |
| 1.2228 × e− 5 | 9.1433 × e− 3 | 29.0708 | Intracerebral calcification on CT scan (human phenotype) | |
| 1.6101 × e− 5 | 1.0702 × e− 2 | 5.6358 | Rhabdomyolysis (human phenotype) | |
| 1.7984 × e− 5 | 1.0758 × e− 2 | 6.2966 | Hypomyelination (human phenotype) | |
| 6.2186 × e− 5 | 3.3818 × e− 2 | 4.7927 | Abnormal formation of myelin sheaths (human phenotype) | |
| 8.7600 × e− 5 | 4.3669 × e− 2 | 5.8403 | Facial muscle weakness, mild (human phenotype) | |
| 1.4132 × e− 6 | 3.1556 × e− 3 | 13.1098 | Gonadoblastoma (disease ontology) | |
| 1.8216 × e− 6 | 2.0338 × e− 3 | 16.9861 | Congenital mesoblastic nephroma (disease ontology) | |
| 4.4680 × e− 6 | 3.3257 × e− 3 | 14.5354 | Phyllodes tumor (disease ontology) | |
| 4.5160 × e− 5 | 2.5211 × e− 2 | 6.4282 | Cherubism (disease ontology) | |
| 5.5084 × e− 5 | 2.4601 × e− 2 | 5.4440 | Sertoli cell-only syndrome (disease ontology) | |
| 8.0341 × e− 5 | 2.9900 × e− 2 | 2.7127 | Mixed cell type cancer (disease ontology) | |
| 4.5886 × e− 5 | 1.2940 × e− 2 | 9.6106 | Trans, trans-farnesyl diphosphate biosynthesis (BioCyc pathway) | |
| 1.8216 × e− 6 | 1.5187 × e− 2 | 16.9861 | TS8_mural trophectoderm (MGI expression) | |
| 7.6734 × e− 6 | 3.1986 × e− 2 | 8.2531 | TS21_perioptic mesenchyme (MGI expression) | |
| 1.0616 × e− 5 | 2.9503 × e− 2 | 12.4855 | TS12_cavities (MGI expression) |
Abbreviations: AGA, appropriate for gestational age; IUGR, intrauterine growth restriction; LGA, large for gestational age; GO, gene ontology; GTP, trimeric G proteins; FDR, false discovery rate; CTD, C-terminal domain; MGI, mouse gene informatics.
aSignificant functional pathways that were enriched in comparisons between IUGR versus AGA, LGA versus AGA, and LGA versus IUGR using GREAT, along with hyper raw P values, hyper FDR Q-values, and hyper fold enrichment values.
STRING analyses of our DMG identify potential interacting partners and enriched functional modules. Our results indeed support those seen in the functional analyses, with biological processes, molecular function, and cellular components involving cellular response to growth factors and protein, small molecule binding and transport, and nervous system development (Table 6).
Table 6.
STRING Protein-Protein Interaction Network Analyses.a
| Comparison | Count In Network | False Discovery Rate | Top Functional Enrichments |
|---|---|---|---|
| IUGR vs AGA | 29 | 1.93d-14 | Homophilic cell adhesion via plasma membrane adhesion molecules (GO Biological Process) |
| 30 | 4.58 × e− 13 | Cell-cell adhesion via plasma-membrane adhesion molecules (GO Biological Process) | |
| 143 | 6.34 × e− 09 | System development (GO Biological Process) | |
| 154 | 5.85 × e− 08 | Multicellular organismal development (GO Biological Process) | |
| 43 | 5.85 × e− 08 | Cell-cell adhesion (GO Biological Process) | |
| 43 | 7.29 × e− 05 | Calcium ion binding (GO Molecular Function) | |
| 152 | 0.000657 | Plasma membrane (GO Cellular Component) | |
| 154 | 0.000657 | Cell periphery (GO Cellular Component) | |
| 176 | 0.00171 | Intrinsic component of membrane (GO Cellular Component) | |
| 68 | 0.00171 | Cell projection (GO Cellular Component) | |
| 169 | 0.0051 | Integral component of membrane (GO Cellular Component) | |
| 11 | 0.00244 | Protein digestion and absorption (KEGG pathways) | |
| 10 | 0.0131 | Pancreatic secretion (KEGG pathways) | |
| 9 | 0.0281 | ECM-receptor interaction (KEGG pathways) | |
| LGA vs AGA | 18 | 0.00292 | Regulation of cellular response to growth factor stimulus (GO Biological Process) |
| 62 | 0.00656 | Nervous system development (GO Biological Process) | |
| 64 | 0.00656 | Regulation of developmental process (GO Biological Process) | |
| 69 | 0.00806 | Regulation of multicellular organismal process (GO Biological Process) | |
| 93 | 0.00873 | System development (GO Biological Process) | |
| 119 | 1.27 × e− 05 | Metal ion binding (GO Molecular function) | |
| 151 | 0.000262 | Ion binding (GO Molecular function) | |
| 30 | 0.00647 | Calcium ion binding (GO Molecular function) | |
| 217 | 0.00692 | Binding (GO Molecular function) | |
| 111 | 0.0338 | Protein binding (GO Molecular function) | |
| 12 | 0.0249 | Cytoplasmic side of plasma membrane (GO cellular component) | |
| 50 | 0.0249 | Cell projection (GO cellular component) | |
| 32 | 0.0249 | Neuron projection (GO cellular component) | |
| 37 | 0.0261 | Neuron part (GO cellular component) | |
| 204 | 0.0262 | Cytoplasm (GO cellular component) | |
| LGA vs IUGR | 27 | 7.68 × e− 13 | Hemophilic cell adhesion via plasma membrane adhesion molecules (GO Biological Process) |
| 26 | 7.41 × e− 10 | Cell-cell adhesion via plasma-membrane adhesion molecules (GO Biological Process) | |
| 149 | 1.22 × e− 07 | Multicellular organismal development (GO Biological Process) | |
| 92 | 2.3 × e− 07 | Anatomical structure morphogenesis (GO Biological Process) | |
| 162 | 2.3 × e− 07 | Developmental process (GO Biological Process) | |
| 46 | 7.38 × e− 07 | Calcium ion binding (GO Molecular function) | |
| 137 | 0.00367 | Metal ion binding (GO Molecular function) | |
| 138 | 0.00412 | Cation binding (GO Molecular function) | |
| 171 | 1.46 × e− 09 | Cell periphery (GO cellular component) | |
| 167 | 3.43 × e− 09 | Plasma membrane (GO cellular component) | |
| 70 | 0.000131 | Cell projection (GO cellular component) | |
| 50 | 0.00111 | Cell junction (GO cellular component) | |
| 84 | 0.00192 | Plasma membrane part (GO cellular component) | |
| 10 | 0.0104 | Protein digestion and absorption (KEGG pathways) |
Abbreviations: AGA, appropriate for gestational age; IUGR, intrauterine growth restriction; LGA, large for gestational age; GO, gene ontology; KEGG, Kyoto encyclopedia of genes and genomes.
aTop functional enrichments in IUGR versus AGA, LGA versus AGA, and LGA versus IUGR comparisons using STRING, along with counts in network and false discovery rate values.
Gene and Protein Expression of DMGs in the Placenta Using GeneCards
Interrogation of the DMGs associated with the top 50 DMRs for each comparison group demonstrated that overall, our DMG are well represented among previously reported mRNA expression studies in the placenta (largely microarrays and SAGE analyses). Using GeneCards, we found that in all comparisons, 81% to 89% of the top ∼5% of DMG have been previously reported with mRNA expression in the placenta. Specifically, in the IUGR versus AGA comparison, 24 of the 27 DMGs associated with the top 50 DMR have been previously reported as having placental mRNA expression (89%).15 Using the same methods, in the LGA versus AGA comparison, 21 of 26 DMGs had been previously reported (81%), and in the LGA versus IUGR comparison, 30 of 34 genes have been described to be expressed in the placenta (88%).15 This finding demonstrates that there is biologic plausibility that our detected DMR/DMG data may result in transcriptomic changes in the placenta.
DNA Methylation and Gene Transcription
Based on the high representation of placental gene expression of our DMGs in the broad, unbiased manner described above, we sought to further examine the relationship between DNA methylation patterns and transcription in specific DMGs. To do this, we performed qRT-PCR on selected DMGs important in metabolic and cardiovascular pathways and immune signaling/inflammation. These pathways were selected to test our hypothesis because the cohorts of interest—IUGR and LGA—have both been shown to be at risk of adult cardiometabolic disease, in part through developmental programming that may occur in the perinatal period,2–6 and inflammation/immune regulation has been shown to be a critical contributor to the pathology of metabolic disease such as diabetes, obesity, and cardiovascular disease. Criteria for selection of these genes were highly significant methylation differences between groups, firstly, and secondly, known clinical significance to human cardiometabolic disease (Table 7). We selected 2 genes from each pathway (metabolic, cardiovascular, immune/inflammatory) to validate gene expression changes—SLC36A1 and PTPRN2 for metabolic disease, CASZ1 and NOS3 for cardiovascular disease, and IL10 and IL32 for inflammation/immune pathways. We were able to demonstrate gene expression differences in 4 of 6 of these genes.
Table 7.
Differentially Methylated Genes in IUGR and LGA Groups Compared to AGA in Metabolic, Cardiovascular, and Inflammatory Pathways.a
| Comparison Groups | Metabolic Genes | Cardiovascular Genes | Inflammatory Genes |
|---|---|---|---|
| IUGR versus AGA | |||
| PCSK1N | PTGIR b | IL10 b | |
| KCNAB2 | NTSR1 b | CD28 | |
| KCND1 | ACE | AZU1 | |
| BLK | NPY | IL3RA | |
| PTPRN2 b | NOS3 | MARCH1 | |
| GK b | CASZ1 b | LRBA | |
| GFPT1 | CD38 | ||
| AGMO | IL32 | ||
| RPH3AL b | |||
| LGA versus AGA | |||
| PTPRN2b | VEGFC | IL10 b | |
| ANKRD23 | PTGIR b | ||
| RIMS1 | CASZ1 b | ||
| GAD2 | CDH13 | ||
| PNPLA7 | NTSR1 b | ||
| GKb | |||
| RPH3ALb | |||
| KCNT1 | |||
| KCNG2 | |||
| BAIAP2 | |||
| LEPR |
Abbreviations: AGA, appropriate for gestational age; IUGR, intrauterine growth restriction; LGA, large for gestational age.
aSelected genes found to be differentially methylated in IUGR and LGA groups compared to controls who have been described as important in human metabolic, cardiovascular, and inflammatory pathways.
bDifferential methylation in both IUGR and LGA compared to AGA.
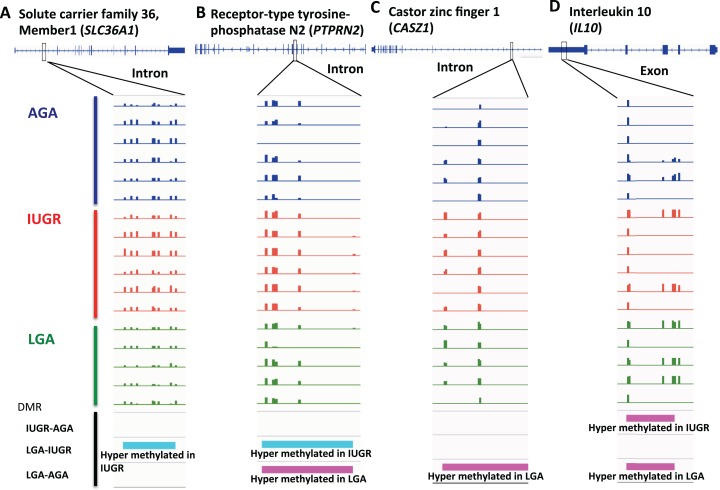
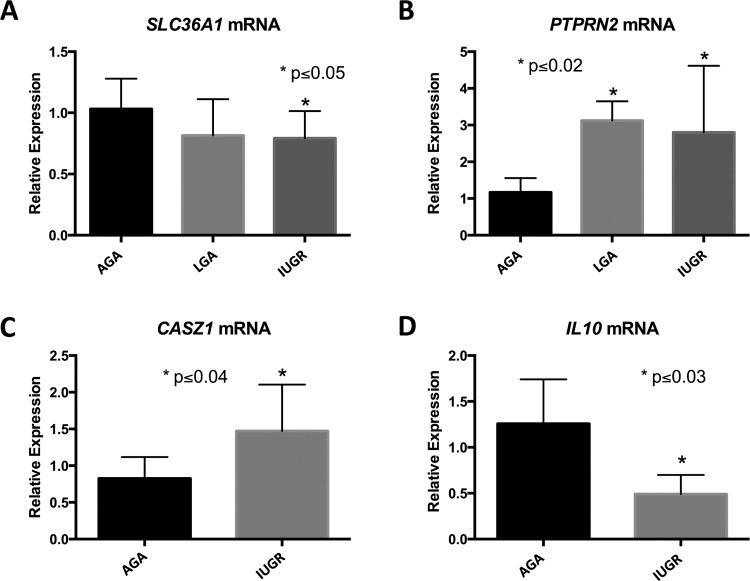
SLC36A1, solute carrier family 36, member 1, encodes a proton-dependent amino acid symporter (also known as PAT1, proton-assisted AAT). Members of this family of proton-assisted amino-acid transporters are important in promoting normal growth via activation of the mammalian target of rapamycin complex 1 (mTORC1) signaling cascade.17 Various groups have demonstrated crosstalk between glucose and fat processing pathways and SLC36A1.18,19 We observed hypermethylation in the intron of SLC36A1 in the IUGR group compared to LGA (Figure 4A). Interestingly, decreased expression is observed in both LGA and IUGR cohorts, though this result is only significant in IUGR (P = .047, ANOVA, n = 9-10/group; see Figure 5A). Thus, in the case of SLC36A1, DNA hypermethylation of an intronic region is associated with decreased expression in IUGR infants.
Figure 4.
Screenshots of DNA methylation tracks in selected genes. A, DMR in SLC36A1 intron is hypermethylated in IUGR comparing with LGA. B, DMR in PTPRN2 intron is hypermethylated in IUGR comparing with LGA, but hypermethylated in LGA comparing with AGA. C, DMR in CASZ1 intron is hypermethylated in LGA comparing with AGA. D, DMR in IL10 exon is hypermethylated in IUGR and LGA, compared to AGA. (magenta: hyper; cyan: hypo). AGA indicates appropriate for gestational age; DMR, differentially methylated region; IL10, interleukin 10; IUGR, intrauterine growth restriction; LGA, large for gestational age.
Figure 5.
q-PCR validation of gene expression. A, SLC36A1 mRNA expression levels in AGA, LGA and IUGR groups. B, PTPRN2 mRNA expression levels in AGA, LGA, and IUGR groups. C, CASZ1 mRNA expression levels in AGA and IUGR groups. D, IL10 mRNA expression levels in AGA and IUGR groups. Data are presented as mean ± SD. All statistical analyses were performed using GraphPad Prism software. Two groups were compared by Student t test with normal distribution, and 3 groups compared by ANOVA with Fisher LSD between groups. Significance was assigned at P ≤ .05. AGA indicates appropriate for gestational age; ANOVA, analysis of variance; DMR, differentially methylated region; IUGR, intrauterine growth restriction; LGA, large for gestational age; q-PCR, quantitative polymerase chain reaction; SD, standard deviation; LSD, least significant difference.
PTPRN2 (protein tyrosine phosphatase, receptor type, N polypeptide 2) encodes a major autoantigen seen in insulin-dependent diabetes mellitus.20,21 We found a total of 12 DMRs within the gene body of PTPRN2 with variable hyper-/hypomethylation patterns in LGA and IUGR, respectively (Figure 4B). Increased gene expression is seen in both LGA and IUGR groups compared to controls (P = .01 and .02, respectively, ANOVA, n = 4-8/group; see Figure 5B). Thus, it appears that gene body methylation of PTPRN2 is a hotspot for DMRs with mixed methylation patterns that are likely to be associated with altered expression in LGA as well as IUGR.
A third gene of interest is CASZ1 (castor zinc finger 1), which encodes a transcription factor that may function as a tumor suppressor. Single-nucleotide polymorphisms in this gene have been associated with blood pressure variation in Asian populations.22,23 We found hypermethylation in an intron of CASZ1 in LGA samples compared to the AGA group (Figure 4C). Increased gene expression is seen in IUGR placentas compared to AGA (P = .03, t test, n = 6-7/group; see Figure 5C). Therefore, intron methylation appears to be correlated with CASZ1 expression.
Lastly, IL10 (interleukin 10) is an important cytokine that promotes immune tolerance in pregnancy.24 Interleukin 10 deficiency coupled with other insults, such as infection or hypoxia in pregnancy, disturbs the balance between anti- and pro-inflammatory factors at the maternal–fetal interface. Resulting ER stress, inflammation, apoptosis, and release of antiangiogenic factors result in poor maternal immune tolerance and perturbed vascular remodeling, which have been associated with adverse pregnancy outcomes.25,26 Interestingly, both LGA and IUGR groups are hypermethylated at the last exon compared to the AGA group (Figure 4D). Interleukin 10 expression is decreased in IUGR compared to AGA (P = .03, t test, n = 4/group; see Figure 5D), which implies decreased immune tolerance in pregnancies complicated by IUGR. This phenomenon of decreased immune tolerance has been shown to be important in the pathophysiology of pregnancy disorders such as preeclampsia, spontaneous abortion, and preterm birth.
Discussion
There is an abundant amount of epidemiologic evidence supporting fetal programming of adult disease. However, the mechanisms underlying these processes are still poorly understood. Studies evaluating epigenetic mechanisms are just beginning to scratch the surface in understanding alteration in disease risk over any individual’s lifetime as well as transmission of disease risk across generations. The present study employs genome-wide DNA methylation analysis in fetal placental tissues associated with altered fetal growth patterns (IUGR and LGA, compared to AGA) to identify novel markers of risk for later cardiometabolic disease. By using placentas sampled from term deliveries carefully selected to limit potential maternal health or environmental exposures on fetal growth, we generated a cumulative picture of how the intrauterine environment affecting fetal growth results in methylation changes affecting genes important for cardiometabolic disease. In addition, the transcriptional profile resulting from these methylation changes that may affect future risk for cardiovascular and metabolic disease was determined.
Our sampling techniques and analytic methods used are either consistent or significantly improved over consensus approaches.27,28 The sampling of the placentas in our project followed several methodological recommendations, including sampling in a consistent location and across multiple sites.28 In addition, we limited our placentas to those taken from term, singleton deliveries to reduce inherent differences in the population being studied. Our methylome analysis followed the guidelines in terms of data reduction and presentation.28 Our data from targeted bisulfite sequencing (RRBS) showed 2 advantages over the common Illumina 450K methylation array: RRBS investigates 4.3M CpG sites, significantly outperforming the 481K CpG sites of the Illumina 450K methylation array, and RRBS provides digital estimates of methylation levels, whereas the array based estimation is based on signal comparison. However, as DMR analysis can be affected by digestion and profiling approaches, we recognize that using RRBS may certainly lead to different interrogation of the methylome and may result in differing results within the separate cohorts as DNA methylation is highly dynamic and may select for alternative transient methylation changes. Our group has also found that the overlap between Illumina 450K array and RRBS sites is quite small, around 5%. Thus, many of the regions identified by one approach will be missed by the other and may explain discrepancies in our data set compared to previously published studies utilizing other methylation analysis methods.29,30
Because our sample size was limited in each group, we decided to use a set of reasonable criteria for defining differential methylation that allowed us to select approximately the top 1% of sites. Despite our best attempts, it is difficult to estimate the true FDRs of these regions, and we acknowledge that they may in fact be quite high. As such, we present these regions as loci that are suggestive of differences between groups and, as a whole, show that the 3 groups do in fact differ in their methylation profiles. Rather than validating each of the individual loci, which is technically demanding, we chose to validate changes in expression among a subset of the genes associated with these loci. It is well established that DNA methylation patterns are associated with gene expression levels, especially in regions defining enhancers and promoters. In fact, the differentially methylated regions we identified are preferentially located within regulatory regions of the genome, in or around CpG islands, as these are known to have important implications on transcriptional regulation.31 In the end, we found that 4 of 6 genes we chose to validate using RTPCR showed significant differential expression, suggesting that the differences in methylation we observed are associated with transcriptional regulation of closely related genes.
The genome-wide pattern of placental methylation in IUGR fetuses and hypomethylation in LGA fetuses is interesting. The global methylation level difference we observe between the groups is within 2% and does not reach statistical significance with a standard t test (Figure 1; Supplementary File 4). However, previous studies have reported global changes in methylation patterns in placentas associated with other pregnancy disorders, such as maternal gestational diabetes.32,33 Our current study supports the growing body of evidence that altered nutrient provision to the fetus affects patterns of methylation across the genome in the placenta, linking the effect of intrauterine environment to regulation of the offspring’s gene expression (Figure 2). Analysis of the patterns suggests that certain areas of the genome are differentially affected by intrauterine environment influencing fetal growth. The pathways that are represented in these areas are specifically enriched for nutrient transfer, metabolism, and hormone synthesis.
We identified 1015, 906, and 1022 DMRs, corresponding to 583, 516, and 567 genes that show significant differential methylation in comparisons between IUGR versus AGA, LGA versus AGA, and LGA versus IUGR. These genes tend to be clustered within specific genomic regions (see Supplementary File 13), supporting the notion that while global methylation levels may not be affected by placental nutrient provision, certain regions may be more susceptible than others. These regions may promote fetal survival and adaptation in the face of suboptimal (as in IUGR) or excessive (as in LGA) nutrient provision, as the associated gene networks identified highlight pathways important in placental function and fetal development (see Table 2).
Major networks identified by multiple pathway and functional analyses highlight the importance of placental cellular structure and cell-to-cell signaling, interaction, assembly, as well as connective tissue development and function. However, network analysis also interestingly identifies methylation differences in genes that are important in the eventual risk of cardiometabolic disease. We found differences in methylation of genes such as growth factors and key regulators of nutrient transport and glucose metabolism, such as TCF7L2, PKLR, PTPRN2, KCNA5, INS, GK, BAIAP2, and AGMO. Although these findings suggest that this epigenomic data set may reflect true pathophysiological processes, further mechanistic studies of these nutrient regulators need to be carried out to establish causal disruptions in fetal growth phenotypes. In addition, several interesting genes that may play a role in endothelial dysfunction, as a precursor for hypertension and cardiovascular disease in the offspring’s later life or subsequent generations, are identified, including, CASZ1, ADD2, CACNA1H, CDH4, NPR3, and NTSR1. It is important to consider that pathway analysis allows exploration of biological mechanisms affected by fetal growth-associated epigenetic variation but does not establish causative roles. Large clinical longitudinal and intervention-based studies would be needed to prove causal relationships and/or provide evidence for programmed risk of disease.
To evaluate whether methylation changes result in biologically important transcriptional changes, we selected specific genes for validation to associate changes in gene methylation with changes in gene expression. Although there were a number of candidate genes that have cardiovascular, metabolic, or immunologic function identified as differentially methylated between IUGR, LGA, and AGA groups, we selected genes with known associations with human disease (Table 7). This was done to highlight the potential clinical implications of these findings and to establish biologic importance to our findings. Specifically, a number of cardiometabolic genes were significantly differentially methylated between groups and merit future study to uncover novel pathways important in programming of human disease. Our results show an overall association (mostly anticorrelation) between DNA methylation and transcription, specifically in SLC36A1, CASZ1, and IL10 genes. It is still unclear how DNA methylation in the gene body affects transcription. However, in recent studies, it has been shown that the patterning of DNA methylation depends critically on the activity of de novo methyltransferases, DNMT3a and b, and it was recently reported that DNMT3b colocalizes with methylated regions. It was also shown that DNMT3b and 5meC are strongly reciprocally correlated with H3K4me3 and positively correlated with H3K36me3.34 These results suggest that actively transcribed genes that are covered with H3K36me3 tend to have higher DNA methylation over the gene body.
There are a number of limitations to this study. DNA isolated from whole placental tissues contains several different cell types, which may limit our ability to find pathways that are altered on a cell-specific basis. Placental chorionic villi contain both trophoblast and mesenchymal cells derived from different embryonic origins. It is known that DNA methylation levels vary between these components and between differing types of trophoblast and mesenchymal cells, but the relative contribution of these cell types to DNA methylation is unknown. The syncytiotrophoblast cell is likely the main contributor to DNA content derived from the term placenta, and isolation of this population via cell sorting techniques is currently difficult and not optimized. The small numbers of individuals per cohort of interest is a limitation as well, although the detection of DNA methylation differences is associated with differential gene expression in 4 of the 6 genes we tested.
Conclusion
In summary, we have identified epigenomic variation and gene expression differences in placental samples associated with pregnancies resulting in AGA, IUGR, and LGA infants. Characterization of these patterns of epigenetic changes supports differences in placental methylation by prenatal growth patterns and birthweight group. Furthermore, pathway analysis suggests that intrauterine environment affecting fetal growth may have a functional impact on processes important in placental function, as well as multiple signaling pathways important in fetal growth and metabolism, cardiovascular disease, and inflammation. Cross-sectional studies, such as this one, offer important insights into the effect of intrauterine environment on fetal tissue. Additionally, they begin to identify candidate genes and pathways that may be helpful in the development of biomarkers signaling altered fetal growth and risk for adult cardiometabolic disease. Future studies must include larger sample sizes to improve power to detect differences and control for false discovery, replication across cohorts, and parallel genetic studies to establish intergenerational risk. Further longitudinal sampling and intervention to determine causation and functional studies toward unraveling the mechanisms by which epigenetic variation can exert its effects are necessary.
Supplementary Material
Acknowledgments
We thank Katie Kemp for extraction of DNA from the human samples.
Authors’ Note: Alison Chu, Matteo Pellegrini, and Sherin U. Devaskar designed research. Pao-Yang Chen, Alison Chu, Amit Ganguly, and Shanthie Thamotharan conducted research. Matteo Pellegrini, Carla Janzen and Sherin U. Devaskar provided essential materials. Pao-Yang Chen, Alison Chu, Wen-Wei Liao, Liudmilla Rubbi, Fei-Man Hsu, Larry Lam, Dennis Montoya, Matteo Pellegrini, and Sherin U. Devaskar analyzed the data. Pao-Yang Chen, Alison Chu, Matteo Pellegrini and Sherin U. Devaskar wrote the article. Sherin U. Devaskar had primary responsibility for final content. All authors read and approved the final manuscript. Pao-Yang Chen and Alison Chu have contributed equally to this work. All work was completed at the University of California–Los Angeles in Los Angeles, California.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by NIH grants no. 1R01GM095656-01A1 (to M.P.) and 1U01HD087221 (to S.U.D.), R01HD089714 (to S.U.D.), K12HD034610 (PI: S.U.D.) and UCLA CTSI KL2TR001882 (PI: Mitchell Wong, MD) (to A.C.), and by grants from Academia Sinica and NHRI, Taiwan (NHRI-EXI03-10324SC) to P.Y.C.
Supplementary Material: Supplementary material is available for this article online.
References
- 1. Maccani MA, Marsit CJ. Epigenetics in the placenta. Am J Reprod Immunol. 2009;62(2):78–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Feil R, Fraga MF. Epigenetics and the environment: emerging patterns and implications. Nat Rev Genet. 2012;13(2):97–109. [DOI] [PubMed] [Google Scholar]
- 3. Manolio TA, Collins FS, Cox NJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461(7265):747–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Godfrey KM, Barker DJ. Fetal programming and adult health. Public Health Nutr. 2001;4(2B):611–624. [DOI] [PubMed] [Google Scholar]
- 5. Curhan GC, Chertow GM, Willett WC, et al. Birth weight and adult hypertension and obesity in women. Circulation. 1996;94(6):1310–1315. [DOI] [PubMed] [Google Scholar]
- 6. McCance DR, Pettitt DJ, Hanson RL, Jacobsson LT, Knowler WC, Bennett PH. Birth weight and non-insulin dependent diabetes: thrifty genotype, thrifty phenotype, or surviving small baby genotype? BMJ. 1994;308(6934):942–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen PY, Ganguly A, Rubbi L, et al. Intrauterine calorie restriction affects placental DNA methylation and gene expression. Physiol Genomics. 2013;45(14):565–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Banister CE, Koestler DC, Maccani MA, Padbury JF, Houseman EA, Marsit CJ. Infant growth restriction is associated with distinct patterns of DNA methylation in human placentas. Epigenetics. 2011;6(7):920–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lillycrop KA, Burdge GC. Environmental challenge, epigenetic plasticity and the induction of altered phenotypes in mammals. Epigenomics. 2014;6(6):623–636. [DOI] [PubMed] [Google Scholar]
- 10. Janzen C, Lei MY, Cho J, Sullivan P, Shin BC, Devaskar SU. Placental glucose transporter 3 (GLUT3) is up-regulated in human pregnancies complicated by late-onset intrauterine growth restriction. Placenta. 2013;34(11):1072–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Smith ZD, Gu H, Bock C, Gnirke A, Meissner A. High-throughput bisulfite sequencing in mammalian genomes. Methods. 2009;48(3):226–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Guo W, Fiziev P, Yan W, et al. BS-Seeker2: a versatile aligning pipeline for bisulfite sequencing data. BMC Genomics. 2013;14(1):774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. McLean CY, Bristor D, Hiller M, et al. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28(5):495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Szklarczyk D, Franceschini A, Wyder S, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43(Database issue):D447–D452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Stelzer G, Rosen N, Plaschkes I, et al. The GeneCards suite: from gene data mining to disease genome sequence analyses. Curr Protoc Bioinformatics. 2016;54:1.30.1–1.30.33. [DOI] [PubMed] [Google Scholar]
- 16. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25(4):402–408. [DOI] [PubMed] [Google Scholar]
- 17. Heublein S, Kazi S, Ogmundsdóttir MH, et al. Proton-assisted amino-acid transporters are conserved regulators of proliferation and amino-acid-dependent mTORC1 activation. Oncogene. 2010;29(28):4068–4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Do TT, Hindlet P, Waligora-Dupriet AJ, et al. Disturbed intestinal nitrogen homeostasis in a mouse model of high-fat diet-induced obesity and glucose intolerance. Am J Physiol Endocrinol Metab. 2014;306(6):E668–E680. [DOI] [PubMed] [Google Scholar]
- 19. Walker DK, Drummond MJ, Dickinson JM, et al. Insulin increases mRNA abundance of the amino acid transporter SLC7A5/LAT1 via an mTORC1-dependent mechanism in skeletal muscle cells. Physiol Rep. 2014;2(3):e00238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schmidli RS, Colman PG, Cui L, et al. Antibodies to the protein tyrosine phosphatases IAR and IA-2 are associated with progression to insulin-dependent diabetes (IDDM) in first-degree relatives at-risk for IDDM. Autoimmunity. 1998;28(1):15–23. [DOI] [PubMed] [Google Scholar]
- 21. Li Q, Borovitskaya AE, DeSilva MG, Wasserfall C, et al. Autoantigens in insulin-dependent diabetes mellitus: molecular cloning and characterization of human IA-2 beta. Proc Assoc Am Physicians. 1997;109(4):429–439. [PubMed] [Google Scholar]
- 22. Takeuchi F, Isono M, Katsuya T, et al. Blood pressure and hypertension are associated with 7 loci in the Japanese population. Circulation. 2010;121(21), 2302–2309. [DOI] [PubMed] [Google Scholar]
- 23. Lu X, Wang L, Lin X, et al. Genome-wide association study in Chinese identifies novel loci for blood pressure and hypertension. Hum Mol Genet. 2015;24(3):865–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cheng SB, Sharma S. Interleukin-10: a pleiotropic regulator in pregnancy. Am J Reprod Immunol. 2015;73(6):487–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Thaxton JE, Sharma S. Interleukin-10: a multi-faceted agent of pregnancy. Am J Reprod Immunol. 2010;63(6):482–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nevers T, Kalkunte S, Sharma S. Uterine regulatory T cells, IL-10 and hypertension. Am J Reprod Immunol. 2011;66(suppl 1):88–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pidsley R, Y Wong CC, Volta M, Lunnon K, Mill J, Schalkwyk LC. A data-driven approach to preprocessing Illumina 450K methylation array data. BMC Genomics. 2013;14:293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hogg K, Price EM, Robinson WP. Improved reporting of DNA methylation data derived from studies of the human placenta. Epigenetics. 2014;9(3):333–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chelbi ST, Mondon F, Jammes H, et al. Expressional and epigenetic alterations of placental serine protease inhibitors: SERPINA3 is a potential marker of preeclampsia. Hypertension. 2007;49(1):76–83. [DOI] [PubMed] [Google Scholar]
- 30. Chelbi ST, Doridot L, Mondon F, et al. Combination of promoter hypomethylation and PDX1 overexpression leads to TBX15 decrease in vascular IUGR placentas. Epigenetics. 2011;6(2):247–255. [DOI] [PubMed] [Google Scholar]
- 31. Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13(7):484–492. [DOI] [PubMed] [Google Scholar]
- 32. Finer S, Mathews C, Lowe R, et al. Maternal gestational diabetes is associated with genome-wide DNA methylation variation in placenta and cord blood of exposed offspring. Hum Mol Genet. 2015;24(11):3021–3029. [DOI] [PubMed] [Google Scholar]
- 33. Ruchat SM, Houde AA, Voisin G, et al. Gestational diabetes mellitus epigenetically affects genes predominantly involved in metabolic diseases. Epigenetics. 2013;8(9):935–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Morselli M, Pastor WA, Montanini B, et al. In vivo targeting of de novo DNA methylation by histone modifications in yeast and mouse. ELife. 2015;4:e06205. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.