Figure 1.
Comparison of Dicer-Independent shRNA Designs and Identification of Potent agshRNAs Targeting VEGF
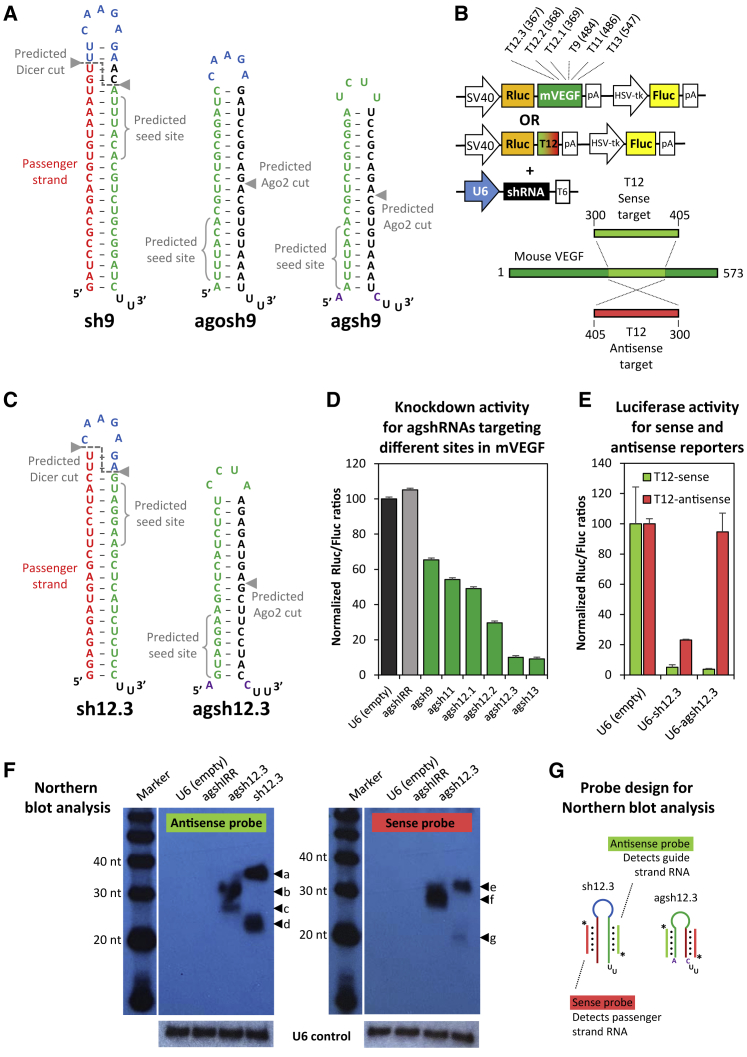
(A) Predicted secondary structure of a previously reported shRNA targeting site 9 in the VEGF gene,26 based on a conventional sense-loop-antisense design (sh9, left side); Ago2-dependent designs, based on the 19/5 agoshRNA structure, as reported by the B. Berkhout lab20 (agosh9, middle part); or the agshRNA design, as described by the Yen group27 (agsh9, right side). Arrows indicate predicted Dicer and Ago2 cleavage sites, and position 2-7 of the guide RNA (seed site) is shown with braces. The guide and passenger strands are depicted in green and red, respectively. The frequently used Brummelkamp loop11 is shown in blue. The invariant unpaired A-C fork, as also present in miR-451, is highlighted in purple. (B) Schematic diagram of the psiCHECK2-based reporter encoding a Renilla luciferase-mVEGF cDNA fusion transcript (used for screening purposes) and the dedicated T12-sense or T12-antisense reporters (used for testing guide and passenger strand activity, respectively), which are used in combination with the various U6-driven shRNA expression plasmids. Name (T9, target site 9) and location (484, 3′ end of target site in the cDNA) of six chosen targets are indicated at the top. The illustration at the bottom shows the 105-nt-long cDNA region of mVEGF (dubbed T12) encompassing site 12, which was cloned into the psiCHECK2-based reporter shown above in sense or antisense orientation (numbers indicate cDNA position). (C) Predicted secondary structure of the conventional sense-loop-antisense sh12.3 (left side) and the Ago2-dependent agsh12.3 (right side) targeting site 12.3 in VEGF. Arrows and colors as indicated in (A). (D) Knockdown activity of agshRNAs designed to target the mVEGF sites, estimated by co-transfection with the mVEGF-fused reporter, as illustrated in the top part of (B). Renilla luciferase (Rluc) and Firefly luciferase (Fluc) activities were measured in relative units of light (RLU). The Rluc:Fluc ratio was normalized to the empty control (black bar) and plotted as the mean of three replicates plus SDs. An irrelevant agshIRR was included as a non-targeting control (gray bar). (E) Guide and passenger strand activity for agsh12.3 and a conventionally designed shRNA predicted to target site 12.3 using the dedicated T12-sense and T12-antisense reporters, respectively (see B). The Rluc:Fluc ratio was normalized to the empty control and plotted as the mean of three replicates plus SDs. (F) Northern blot analysis of small RNA from transfected HEK293 cells using probes detecting guide strand RNA (antisense probe, autoradiogram on the left side) or passenger strand RNA (sense probe, autoradiogram on the right side). Size of 20-, 30-, and 40-nt bands of the RNA decade marker is indicated, and agsh12.3- and sh12.3-specific bands are marked by arrows and labeled a–g. A cropped image with detection of the native U6 snRNA band (loading control) is shown below. (G) Diagram indicating probe specificity. Antisense probe, light green; sense probe, light red; Fluc, Firefly luciferase; HSV-tk, Herpes simplex virus thymidine kinase promoter; mVEGF, murine vascular endothelial growth factor; pA, polyadenylation signal; Rluc, Renilla luciferase; shRNA, short hairpin RNA; SV40, simian virus 40 promoter; T6, T-rich Pol III termination signal; U6, human U6 snRNA promoter.