Abstract
Molecularly imprinted polymers (MIPs) are high affinity robust synthetic receptors, which can be optimally synthesized and manufactured more economically than their biological equivalents (i.e. antibody). In MIPs production, rational design based on molecular modeling is a commonly employed technique. This mostly aids in (i) virtual screening of functional monomers (FMs), (ii) optimization of monomer-template ratio, and (iii) selectivity analysis. We present MIRATE, an integrated science gateway for the intelligent design of MIPs. By combining and adapting multiple state-of-the-art bioinformatics tools into automated and innovative pipelines, MIRATE guides the user through the entire process of MIPs’ design. The platform allows the user to fully customize each stage involved in the MIPs’ design, with the main goal to support the synthesis in the wet-laboratory. Availability: MIRATE is freely accessible with no login requirement at http://mirate.di.univr.it/. All major browsers are supported.
Keywords: docking, molecularly imprinted polymers, science gateway
1. Introduction
Molecularly imprinted polymers (MIPs) have been involved in a huge number of processes, boasting many hundreds of articles published annually in this area ([1] and references within). MIPs are frequently used in chromatography, sensors and assays, and they are considered as the cost-effective substitutes for biological macromolecules in several research areas and practical applications [2]. A molecularly imprinted polymer can be defined as the process of template-induced formation of specific recognition sites (binding or catalytic) in a material where the template directs the positioning and orientation of the material’s structural components by a self-assembling mechanism. In a non-covalent molecular imprinting, for a template molecule (or target), appropriate functional monomers (FMs) are chosen and allowed to form a self-assembly construct. By co-polymerization with a cross-linking molecule, a polymer network is formed in which the self-assembly is set. Thereby, the position and the spatial conformation of the monomers are constructed according to the template. The embedded template can then be extracted and rebound to the molecularly imprinted polymer. The targets are usually small compounds or appropriate binding epitopes [2].
Recent improvements in the synthesis of molecularly imprinted nanoparticles [3] combined with their stability, low cost preparation of these imprinted materials [4] and the efficacy in antimicrobial, antiviral and anticancer therapy [5] have led the MIPs to be recognized as “plastic antibody” [6].
In recent years, computational techniques have been advanced and exploited in the optimization of polymer composition to rationally design high affinity synthetic receptors by using the molecular imprinting technique [7]. One of the most famous molecular mechanics/molecular dynamics protocol for MIPs’ rational design, has been designed by Chianella and collaborators [8] and runs over the Sybyl modeling package of programs (Tripos Inc., St. Louis, MO, USA). Here, we present MIRATE, a new open-source science gateway as alternative to the previous cited patented protocol. MIRATE stems from the desire to find a free alternative computational method to the Chianella’s protocol that relies on purchasing Sybyl. The combination of several state-of-the-art tools in a single publicly available science gateway is MIRATE’s innovation. Nevertheless, MIRATE provides a unique environment to MIPs design and allows users to intervene in the process, preventing it from becoming a fully-automatic black-box.
2. Related Work
Computational design has become a common procedure in the manufacture of MIPs, and it has allowed important improvements in functional monomer selection, calculation of monomers-template ratio and selectivity analysis. As extensively presented by Cowen and collaborators [7], several studies used computational techniques for rational MIPs design (see Table 1, [7] and references within it for more details). Broadly speaking, the used computational techniques can be clustered based on the technique employed (e.g. monomer selection, study of the electronic structure of the template and monomers) and by the used algorithms: those span from molecular mechanics/molecular dynamics to quantum mechanics e.g. semi-empirical, ab initio (Hartree-Fock, Møller–Plesset, etc.) and Density functional theory [9]. The most employed quantum mechanics techniques for MIPs design are based on the use of density functional theory and the hybrid functional B3LYP [10]. Indeed, the combination of them bring together the accuracy of the quantum mechanical calculations with the effectiveness of ab initio method for describing the electronic structure under investigation. Thus, QM methods are commonly used for studying the monomer template interactions in order to investigate the appropriate pre-polymerisation mixture for high affinity and selectivity MIPs [11], [12], [13]. These kind of calculations, although extremely accurate, are very expensive from a computational point of view. In the other hand, molecular mechanics and molecular dynamics simulations are methods that are used for modeling multi-component systems and thus can model many interactions [14]. With these techniques, the screening of several FMs become a feasible task by minimization of energies and optimization of geometries [15]. One of the most used molecular mechanics and molecular dynamics simulations protocol for MIPs design, has been developed by some of us (S.P. polymer group) [8] and it includes the application of the LeapFrog program [16]. Although this protocol is widely used in both academic and industrial fields with successful results [17], a free open access protocol able to produces good predictions in a short period of time is needed. From the best of our knowledge, MIRATE is the first open-source science gateway in molecularly imprinted polymers field. In this context, our publicly accessible science gateway, MIRATE, was developed as a novel alternative for the rational design of MIPs. Our science gateway, offers and combines the predictions of the majority of the state-of-the-art software (such as HADDOCK, Autodock, Gromacs), which have been widely tested in several applications published in papers of high impact. This science gateway stems from the desire to find a free alternative computational method to the standard protocol that relies on purchasing Sybyl [8]. MIRATE, compared with the widely used and tested Sybyl method, has shown very good semblance and sometimes improved the Sybyl results (see Section 4). The predictive nature of MIRATE could result in a range of polymer materials with superior affinity, and selectivity of what is currently being used.
3. Workflow and Architecture
In the last few years we have been developing state-of-the-art, free, in silico molecular modeling techniques [18], [19], [20], [21] to optimize the polymer compositions and to design high affinity synthetic receptors based on molecular imprinting. The MIRATE science gateway aims at providing an efficient and user-friendly solution to apply and to combine all these approaches for the rational design of MIPs through a publicly accessible web-based platform.
Given the most used FMs and the molecular template, MIRATE suggests the putatively most affine FMs and the optimal stoichiometric ratio.
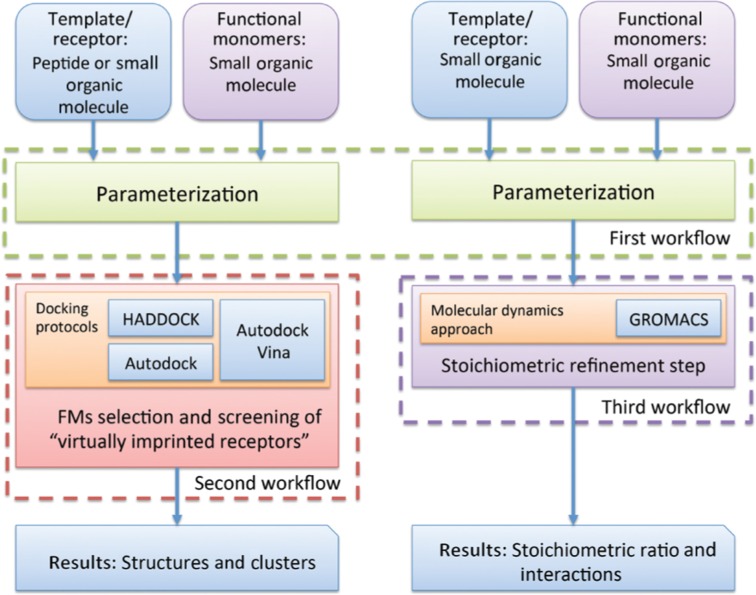
The science gateway provides three different workflows for MIPs’ design (see Figure 1).
Figure 1:
The MIRATE workflows.
The parametrization workflow. This workflow offers the possibility to parametrise the input molecules (template/“virtually imprinted receptor” and FMs) in several ways by using state-of-the-art approaches. Therefore, for instance, the users are driven by a simplified version of the ACPYPE software program [22], which offers several methods to assign charges to small organic molecules. Additionally, MIRATE gives the opportunity to expert users to upload partial charges already precalculated by another program/server such as R.E.D.-III [23] (see Supplementary Material S1.1 for more details about all parametrization protocols implemented in MIRATE).
The second workflow implements two tasks: (i) FMs selection and (ii) screening of “virtually imprinted receptors” for rebinding the molecular template [20], i.e. selectivity analysis. This workflow is based on three well-established docking protocols: (1) the knowledge-based HADDOCK [24], [25] provided through GOMoDo server [26], (2) the Autodock [27] and (3) the Autodock Vina [28] (see Supplementary Material S1.2). MIRATE also includes a virtual library containing the most commonly used parameterized FMs. It enables both, the FMs screening against a template and the “virtually imprinted receptors” screening against the molecular template. Then, the complexes are ranked by binding energy/docking score. Both the docking complexes and clusters are provided as well. The docking information is important for the wet-lab candidates preparation. Indeed, the success of the imprinting procedure is related to the formation of a strong monomer-template complex.
The third workflow relies on a stoichiometric refinement step, which aims at calculating the ideal template/monomer stoichiometric ratio by using a novel molecular dynamics method based on GROMACS [29] (see Supplementary Material S1.3).
For the analysis of the results, the platform provides information regarding the monomers-template interactions (hydrogen bonds) and the number of FMs interacting with the template. The user can determine the type and quantity of FMs participating in the complex, and thus, extracting the ratio of the template and monomers for the polymer synthesis. As developed for the MIPs’ rational design, MIRATE supports molecular templates in input, such as simple short peptides or small organic molecules. Indeed, in the pipeline based on HADDOCK, MIRATE handles short peptides with plain standard aminoacids, free of auxiliary ions, ligands, and with up to one chain.
MIRATE carefully checks all the inputs uploaded by users (e.g. the input peptide in terms of aminoacids and atom type) in order to prevent any issue that could compromise the results.
MIRATE includes a complete documentation and step-by-step tutorials.
3.1. Overview of the MIRATE HW/SW Architecture
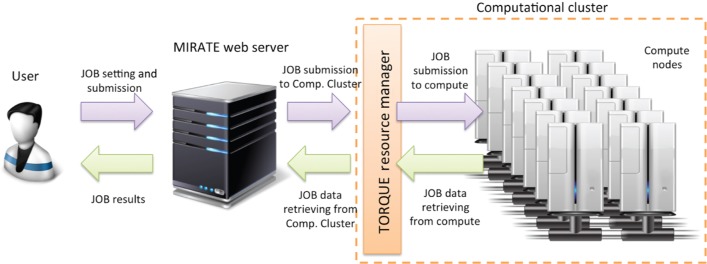
MIRATE provides a whole series of facilities to perform molecular docking and refinement, supporting both peptides and organic molecules. MIRATE does not require any specific library or adjustment to be used, thus allowing users to focus only on simulation and execution. The MIRATE user interface (UI) relies on the latest web technologies, such as HTML5, JQuery [30] and Twitter Bootstrap [31], aiming to increase user experience. The UI is fully responsive and interactive, adapting its content to be visualized through any desktop as well as mobile devices. In order to minimize data exchange between client and server, the platform relies on a custom framework written in PHP language according to the model-view-controller design pattern [32]. The whole MIRATE science gateway is organized into two main blocks: a web interface designed to collect all submitted user jobs and a computational cluster designed for intense computation. The submitted jobs are collected in a dedicated batch queue and processed on a high-performance computer cluster (see Figure 2), using custom workers. A worker (job collector and submitter) is designed to create, and later submit, a new job according to a batch queue. A second worker (data retriever) is designed to retrieve, periodically, the job execution status, updating its information accordingly.
Figure 2:
Overview of the MIRATE SW/HW architecture.
The local cluster is equipped with TORQUE/PBS Resource Manager (http://www.adaptivecomputing.com/products/open-source/torque/), used to distribute and control the batch jobs. TORQUE/PBS assigns each submitted job to each compute node (node of the cluster). In such a way, each computation is executed in a separate environment. In such a way, MIRATE is relieved from intensive computational tasks, providing a reactive system.
4. Method Validation
In order to determine the accuracy and reliability of MIRATE predictions we compared MIRATE with the only one established protocol which employs a Sybyl-based screening model and is already extensively tested and applied in the literature. As the monomers used for the tests possess different physicochemical properties, therefore our virtual library contains acidic monomers: acrylamido-2-methyl-1-propanesulfonic acid (AMPSA), acrylic acid (AA), itaconic acid (IA), methacrylic acid (MAA) and trifluoromethylacrylic acid (TFMAA); basic monomers: allylamine, 1-vinylimidazole (VI), 2-vinylpyridine (2-VP), 4-vinylpyridine (4-VP) and N,N-diethylamino ethyl methacrylate (DEAEM) and neutral monomers: acrolein, acrylamide, acrylonitrile, m-divinylbenzene, p-divinylbenzene, ethylene glycol dimethacrylate (EGDMA), 2-hydroxyethyl methacrylate (HEM), N,N -methylenebisacrylamide (MBAA) and styrene. MIRATE was found to match that of the Sybyl model with a 28.8% error in ranking from a sample size of 114 combinations (see Tables 2.1–2.3 in Supplementary Material). MIRATE shows very good semblance with Sybyl and for two (NK11 and AK9) out of three templates predicted itaconic acid as the best monomer in agreement with the experimental data while Sybyl predicted itaconic acid only as the second-best monomer. We have analyzed also the performance of the docking programs that showed a high agreement with experimental results (see Supplementary Material S2.2 for Autodock/Vina’s explanation).
5. Application Cases
We applied MIRATE to evaluate the efficiency and the automation of the computational workflows for several MIPs application cases [18], [19], [20], [21]. We exploited the first and the second workflow in two of them, by which we synthetized high-affinity synthetic receptors [18], [19]. These included a high-affinity receptor for the iron-regulating hormone Hepcidin-25 [18], which is a significant marker for cardiovascular risk, and three receptors for the cardiac-troponin-I peptides (NITEIADLTQK, NIDALGMEGR and AYATEPHAK), which are important markers for cardiovascular risk [19].
We also applied the first and second workflow to evaluate the MIPs’ rebinding ability (i.e. selectivity analysis), by taking into account the cumulative steric effect of multiple FMs within the receptor site [20]. Indeed, the methodology, here, fully implemented in MIRATE, has been successfully applied in three cases, either confirming the experimental data available and aiding the synthesis of more selective MIPs. The application cases of this methodology were: (1) the detection of tetrahydrocannibinol using a Catechin–hydrate MIP with acrylamide as monomers; (2) the detection of Bisphenol A and Bisphenol F using a Bisphenol A MIP with 4-vinylpyridine as the monomer and (3) the detection of hypoxanthine by a selection of theophylline MIPs.
By using the first and third workflows, computational results also allowed us to synthesize and validate in a wet-lab an efficient MIP for melamine, a heterocyclic triazine that is very important in the “food safety” area [21].
6. Discussion
This unique publicly free available science gateway implements a novel protocol for the rational design of MIPs for practical applications. The science gateway allows a strong user intervention at different stages offering the user, the choice between the parameter for MIPs modeling and default parameters. However, the expert user can always modify the default parameters along the pipeline and introduce experimental information within the server limitations. Although we recommend preferential routes to be followed along the web server, according to some points e.g. the type molecules uploaded, the users’ goals and our case-studies; we always give maximum freedom to the user. Indeed, the protocol has been shown in several cases [18], [19], [20], [21] to be useful to analyse mainly the monomer-template and template-receptor interactions in a physiological water solvent, and to offer a reliable (within the limitations of the methods) prediction, not only on the monomers to be used but also on the concentration ratio. MIRATE is thus the first publicly available computational protocol aimed at the rational design of MIPs. This new free science gateway offers the possibility of performing fast screening of adsorbents and it is suitable for resolving particular separation/diagnostic problems. The main beneficiaries of the present work will be the scientific community as well as pharmaceutical and fine chemicals companies. Indeed, MIRATE, will pave the way for many experimental labs, using brute-force trial/error approaches, leading them to use this innovative computer-assisted way for synthetic antibodies design.
Supporting Information
Supplemental Material
The online version of this article offers supplementary material (DOI: jib-2017-0075).
Conflict of interest statement
Authors state no conflict of interest. All authors have read the journal’s Publication ethics and publication malpractice statement available at the journal’s website and hereby confirm that they comply with all its parts applicable to the present scientific work.
References
- [1].Whitcombe MJ, Kirsch N, Nicholls IA. Molecular imprinting science and technology: a survey of the literature for the years 2004–2011. J Mol Recognit. 2014;27:297–401. doi: 10.1002/jmr.2347. [DOI] [PubMed] [Google Scholar]
- [2].Lu CH, Zhang Y, Tang SF, Fang ZB, Yang HH, Chen X. et al. Sensing HIV related protein using epitope imprinted hydrophilic polymer coated quartz crystal microbalance. Biosens Bioelectron. 2012;31:439–44. doi: 10.1016/j.bios.2011.11.008. [DOI] [PubMed] [Google Scholar]
- [3].Canfarotta F, Poma A, Guerreiro A, Piletsky S. Solid-phase synthesis of molecularly imprinted nanoparticles. Nat Protoc. 2016;11:443–55. doi: 10.1038/nprot.2016.030. [DOI] [PubMed] [Google Scholar]
- [4].Poma A, Guerreiro A, Whitcombe MJ, Piletska EV, Turner AP, Piletsky SA. Solid-phase synthesis of molecularly imprinted polymer nanoparticles with a reusable template–“Plastic Antibodies”. Adv Funct Mater. 2013;23:2821–7. doi: 10.1002/adfm.201202397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Puoci F, Cirillo G, Curcio M, Parisi OI, Iemma F, Picci N. Molecularly imprinted polymers in drug delivery: state of art and future perspectives. Expert Opin Drug Deliv. 2011;8:1379–93. doi: 10.1517/17425247.2011.609166. [DOI] [PubMed] [Google Scholar]
- [6].Hoshino Y, Shea KJ. The evolution of plastic antibodies. J Mat Chem. 2011;21:3517–21. [Google Scholar]
- [7].Cowen T, Karim K, Piletsky S. Computational approaches in the design of synthetic receptors–A review. Analytica Chimica Acta. 2016;936:62–74. doi: 10.1016/j.aca.2016.07.027. [DOI] [PubMed] [Google Scholar]
- [8].Chianella I, Lotierzo M, Piletsky SA, Tothill IE, Chen B, Karim K. et al. Rational design of a polymer specific for microcystin-LR using a computational approach. Anal Chem. 2002;74:1288–93. doi: 10.1021/ac010840b. [DOI] [PubMed] [Google Scholar]
- [9].Nicholls IA, Andersson HS, Charlton C, Henschel H, Karlsson BC, Karlsson JG. et al. Theoretical and computational strategies for rational molecularly imprinted polymer design. Biosens Bioelectron. 2009;25:543–52. doi: 10.1016/j.bios.2009.03.038. [DOI] [PubMed] [Google Scholar]
- [10].Gao B, He XP, Jiang Y, Wei JT, Suo H, Zhao C. Computational simulation and preparation of fluorescent magnetic molecularly imprinted silica nanospheres for ciprofloxacin or norfloxacin sensing. J Sep Sci. 2014;37:3753–9. doi: 10.1002/jssc.201401014. [DOI] [PubMed] [Google Scholar]
- [11].Khan MS, Pal S, Krupadam RJ. Computational strategies for understanding the nature of interaction in dioxin imprinted nanoporous trappers. J Mol Recognit. 2015;28:427–37. doi: 10.1002/jmr.2459. [DOI] [PubMed] [Google Scholar]
- [12].Wang Y, Liu JB, Tang SS, Jin RF, Chang HB. Preparation of melamine molecular imprinted polymer by computer aided design. Chem J Chin Univ Chin. 2015;36:945–54. doi: 10.1002/jssc.201500375. [DOI] [PubMed] [Google Scholar]
- [13].Jun-Bo L, Yang S, Shan-Shan T, Rui-Fa J. Theoretical and experimental research on the self-assembled system of molecularly imprinted polymers formed by salbutamol and methacrylic acid. J Sep Sci. 2015;38:1065–71. doi: 10.1002/jssc.201401309. [DOI] [PubMed] [Google Scholar]
- [14].Uzuriaga-Sanchez RJ, Khan S, Wong A, Picasso G, Pividori MI, Sotomayor MD. Magnetically separable polymer (Mag-MIP) for selective analysis of biotin in food samples. Food Chem. 2016;190:460–7. doi: 10.1016/j.foodchem.2015.05.129. [DOI] [PubMed] [Google Scholar]
- [15].Mistry J, Guerreiro A, Moczko E, Piletska E, Karim K. Piletsky, S. Analysis of cooperative interactions in molecularly imprinted polymer nanoparticles. Mol Imprint. 2015;2:83–92. [Google Scholar]
- [16].Abdin MJ, Altintas Z, Tothill IE. In silico designed nanoMIP based optical sensor for endotoxins monitoring. Biosens Bioelectron. 2015;67:177–83. doi: 10.1016/j.bios.2014.08.009. [DOI] [PubMed] [Google Scholar]
- [17].Piletsky SA, Karim K, Piletska EV, Day CJ, Freebairn KW, Legge C. et al. Recognition of ephedrine enantiomers by molecularly imprinted polymers designed using a computational approach. Analyst. 2001;126:1826–30. [Google Scholar]
- [18].Cenci L, Anesi A, Busato M, Guella G. Bossi, AM. Molecularly imprinted polymers coupled to matrix assisted laser desorption ionization mass spectrometry for femtomoles detection of cardiac troponin I peptides. J Mol Recognit. 2016;29:41–50. doi: 10.1002/jmr.2494. [DOI] [PubMed] [Google Scholar]
- [19].Cenci L, Andreetto E, Vestri A, Bovi M, Barozzi M, Iacob E. et al. Surface plasmon resonance based on molecularly imprinted nanoparticles for the picomolar detection of the iron regulating hormone Hepcidin-25. J Nanobiotechnol. 2015;13:51. doi: 10.1186/s12951-015-0115-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Bates F, Cela-Pérez MC, Karim K, Piletsky S, López-Vilariño JM. Virtual screening of receptor sites for molecularly imprinted polymers. Macromol Biosci. 2016;16:1170–4. doi: 10.1002/mabi.201500461. [DOI] [PubMed] [Google Scholar]
- [21].Bates F, Busato M, Piletska E, Whitcombe MJ, Karim K, Guerreiro A. et al. Computational design of molecularly imprinted polymer for direct detection of melamine in milk. Separation Science and Technology. 2017;52(8):1441–53. [Google Scholar]
- [22].da Silva AW, Vranken WF. ACPYPE-Antechamber python parser interface. BMC Res Notes. 2012;5:367. doi: 10.1186/1756-0500-5-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Dupradeau FY, Pigache A, Zaffran T, Savineau C, Lelong R, Grivel N. et al. The REd. Tools: Advances in RESP and ESP charge derivation and force field library building. Phys Chem Chem Phys. 2010;12:7821–39. doi: 10.1039/c0cp00111b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Dominguez C, Boelens R, Bonvin AM. HADDOCK: a protein-protein docking approach based on biochemical or biophysical information. J Am Chem Soc. 2003;125:1731–7. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- [25].van Zundert GC, Rodrigues JP, Trellet M, Schmitz C, Kastritis PL, Karaca E. et al. The HADDOCK2.2 Web Server: user-friendly integrative modeling of biomolecular complexes. J Mol Biol. 2016;428:720–5. doi: 10.1016/j.jmb.2015.09.014. [DOI] [PubMed] [Google Scholar]
- [26].Sandal M, Duy TP, Cona M, Zung H, Carloni P, Musiani F. et al. GOMoDo: a GPCRs online modeling and docking webserver. PLoS One. 2013;8:e74092. doi: 10.1371/journal.pone.0074092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS. et al. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem. 2009;30:2785–91. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–61. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Pronk S, Páll S, Schulz R, Larsson P, Bjelkmar P, Apostolov R. et al. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics. 2013;29:845–54. doi: 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Bear B, Katz Y. jQuery Foundation. jQuery. 2008. http://jquery.com. [Google Scholar]
- [31].Otto M, Thornton J. Twitter Bootstrap. 2013. https://getbootstrap.com/2.0.4/ [Google Scholar]
- [32].Leff A, Rayfield JT. Web-application development using the Model/View/Controller design pattern; Proceedings Fifth IEEE International Enterprise Distributed Object Computing Conference; 2001. pp. 118–127. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.