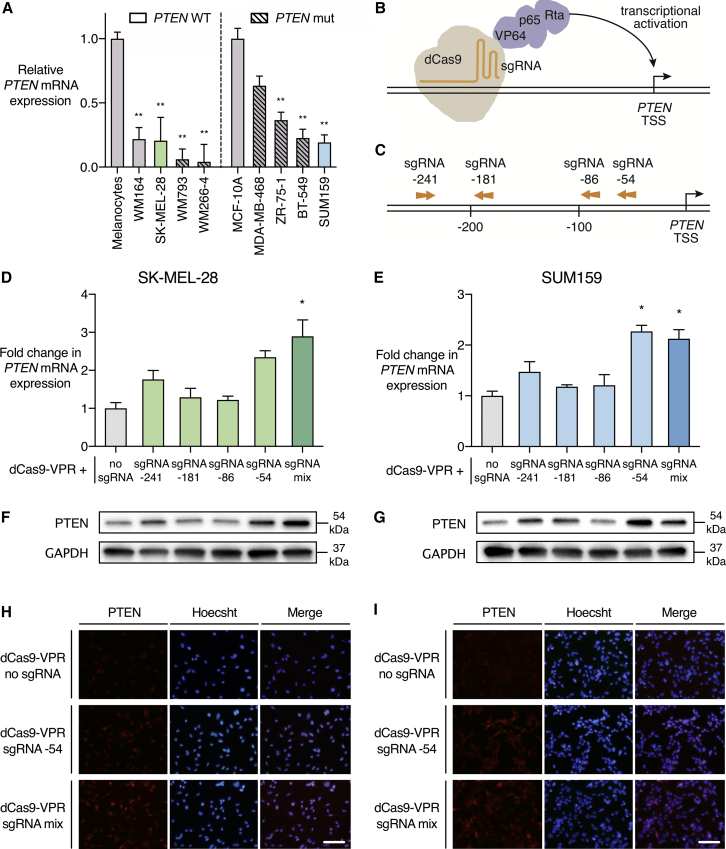
Figure 1.
PTEN Is Activated by the dCas9-VPR System in SK-MEL-28 Melanoma and SUM159 TNBC Cell Lines
(A) PTEN mRNA expression in a panel of BRAF mutant melanoma and TNBC cell lines. PTEN wild-type (WT) or mutant (mut) status is indicated with clear and hatched bars, respectively. Data are presented as PTEN mRNA expression level relative to normal human melanocytes or normal-like immortalized breast epithelial cells (MCF-10A). SK-MEL-28 and SUM159 were selected as cell line models with wild-type but transcriptionally downregulated PTEN. *p < 0.05, **p < 0.01; n = 3; error bars show SEM. (B) Schematic representation of the sgRNA-dCas9-VPR complex. dCas9 is in direct C-terminal fusion with transactivators VP64, p65, and Rta. sgRNAs bind upstream of the PTEN transcription start site (TSS) to activate gene expression. (C) sgRNA target sites in the PTEN proximal promoter region. sgRNA numbering refers to the distance in base pairs from the TSS of PTEN mRNA transcript variant 1. Arrows indicate whether the sgRNA targets the forward or reverse DNA strand. (D–I) dCas9-VPR was stably expressed in SK-MEL-28 and SUM159 cell lines with no sgRNA, individual sgRNAs targeting the PTEN proximal promoter, or a mix of all four PTEN-targeting sgRNAs. (D and E) Fold change in PTEN mRNA expression in qRT-PCR relative to dCas9-VPR with no sgRNA in SK-MEL-28 (D) and SUM159 (E). *p < 0.05, **p < 0.01; n = 3; error bars show SEM. (F and G) Immunoblot of PTEN and GAPDH in SK-MEL-28 (F) and SUM159 (G) cell lines. Conditions correspond to qRT-PCR data labeled above. (H and I) Immunofluorescence of PTEN and Hoechst-stained cell nuclei in SK-MEL-28 (H) and SUM159 (I) cell lines. Scale bars represent 100 μm.