Figure 2.
PTEN Activation by dCas9-VPR Does Not Affect the Expression of Predicted Off-Target Genes
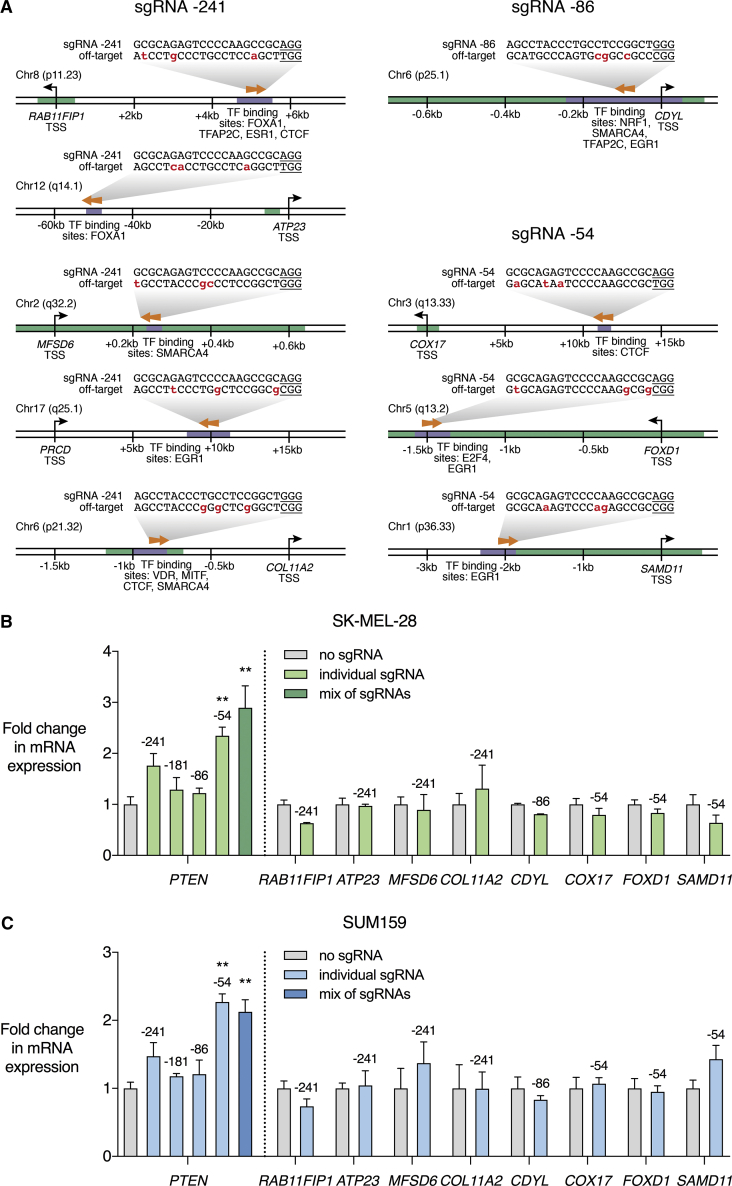
(A) The program Cas-OFFinder was used to identify genomic sequences with 3 mismatches or less to the cognate sequence of any of the 4 PTEN-targeting sgRNAs and upstream of the S. pyogenes NGG protospacer-adjacent motif (PAM). Of the identified off-target sites, nine sites were found in proximity to regulatory elements and were associated with greater potential to modulate gene expression. The potential off-target genes were RAB11FIP1, ATP23, MFSD6, PRCD, COL11A2, CDYL, COX17, FOXD1, and SAMD11. Mismatches between cognate and off-target sequences are highlighted in red and PAM sequences are underlined. Arrows indicate whether the sgRNA targets the forward or reverse DNA strand at the potential off-target binding site. Numbering on the DNA indicates the distance from the transcription start site (TSS) of the indicated gene. Green-shaded regions of DNA represent CpG islands annotated in the UCSC Genome Browser. Purple-shaded regions of DNA represent the location of transcription factor (TF)-binding sites according to literature-curated PAZAR and JASPAR databases. (B and C) dCas9-VPR was stably expressed in SK-MEL-28 (B) and SUM159 (C) cell lines with no sgRNA, individual PTEN sgRNAs, or a mix of all four sgRNAs targeting the PTEN proximal promoter. Data are shown as fold change in mRNA expression in qRT-PCR relative to dCas9-VPR with no sgRNA, for PTEN and each of the nine potential off-target genes. Numbers above the bars indicate the relevant PTEN-targeting sgRNA. There was no significant effect on mRNA expression of any of the predicted off-target genes. Data for PRCD are not shown, as PRCD expression was not reliably detected in either control or sgRNA-treated samples. *p < 0.05; n = 3; error bars show SEM.