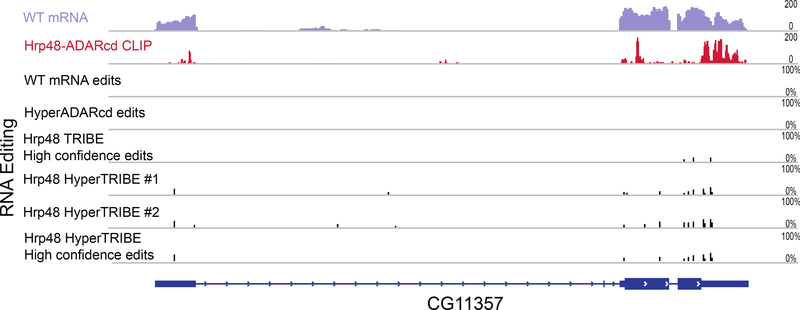
Figure 5. HyperTRIBE fusion protein reproducibly edits more sites compared to TRIBE and has a higher overlap with CLIP signals.
The top two panels show wild type mRNA expression and a CLIP profile for a sample gene, CG11357. The panels directly below show (from top to bottom): RNA editing tracks from wild type cells, cells with HyperADARcd alone, high confidence sites from Hrp48 TRIBE, Hrp48 HyperTRIBE replicate 1 and replicate 2, which are followed by high confidence editing sites from Hrp48 HyperTRIBE at the bottom. The HyperTRIBE data were generated by expressing Hrp48 HyperTRIBE with the elav-geneswitch-Gal4 driver and manually sorting for eGFP-positive cells. The criteria of 20 reads and 10% editing percentage were enforced to select for these editing sites. The black bars in the editing track indicate editing events and the height of the bars shows editing percentage at that site. Editing sites reproduced in at least two replicates with no failures are designated high confidence sites.